
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,993,433 – 21,993,579 |
| Length | 146 |
| Max. P | 0.998816 |

| Location | 21,993,433 – 21,993,552 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.18 |
| Mean single sequence MFE | -25.86 |
| Consensus MFE | -19.77 |
| Energy contribution | -19.68 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21993433 119 - 27905053 UUGGGUUUCCGCUUUCCCCUUGGGCACUUAAACACCUACUUAAGUGCUCAGGCAGAAUUUUCACAAAUUCCUCGUAAAAUUUUAAUUUCAUCACGCAUAUGCAGGAUUUAAGAUAGACU ..(((..........)))(((((((((((((........)))))))))))))..((((((....))))))...((...(((((((....(((..((....))..))))))))))..)). ( -26.10) >DroSim_CAF1 29730 119 - 1 UUGGUUUUCCGUUUUCCCCUUGGGCACUUAAACACCUACUUAAGUGCUCAGGCAGAAUUUUCACAAAUUCCUCGUAAAAUUUUAAUUUCAUCACGCAUAUGCAGGGCUUAAGGUAGAGU ....((((((.....((((((((((((((((........))))))))))))).((((((((.((.........))))))))))...........((....)).))).....)).)))). ( -26.30) >DroEre_CAF1 35393 119 - 1 UUGGCUCUCCCUUUUCCCCUUCGGCACUUAAACACCUACUUAAGUGCUCAGGCAGAAUUUUGACAAAUUCCUCGUAAAAUUUUAAUUUCAUCCAGCGUAUGCAAGGCUCCAGGUGGAGU ...((((..(((.....((((.(((((((((........)))))))))...(((((((((....))))))..(((...................)))..)))))))....)))..)))) ( -25.51) >DroYak_CAF1 34021 119 - 1 UUGGCCCACCGUUUUCCCCUUGGGCACUUAAACACCUACUUAAGUGCUCAGGCAGAAUUUUGACAAACUCCUCGUAAAAUUUUAAUUUCAUCACGCACAUGCAGGGCUAAAGUUAAAAA (((((((...((((((..(((((((((((((........)))))))))))))..)))....)))..............................((....)).)))))))......... ( -32.60) >DroAna_CAF1 39735 101 - 1 UUGG-------UUUUCCCCUCGGGCACUUAAACACCUACUUAAGUGCUUAAGCAAAAUUUUCGCACAUUCUCUGUAAAAUUUUAAUUUCCUCACGCACCUGCA---CUGAA-------- (..(-------(......((..(((((((((........)))))))))..)).((((((((.(((.......)))))))))))...........((....)))---)..).-------- ( -18.80) >consensus UUGGCUCUCCGUUUUCCCCUUGGGCACUUAAACACCUACUUAAGUGCUCAGGCAGAAUUUUCACAAAUUCCUCGUAAAAUUUUAAUUUCAUCACGCAUAUGCAGGGCUUAAGGUAGAGU ..(((((...........(((((((((((((........))))))))))))).((((((((.((.........))))))))))...........((....)).)))))........... (-19.77 = -19.68 + -0.09)


| Location | 21,993,473 – 21,993,579 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.81 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -21.82 |
| Energy contribution | -21.78 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21993473 106 - 27905053 CUU-------------AAACAUUACCAUCAACUUUCGGUGUUGGGUUUCCGCUUUCCCCUUGGGCACUUAAACACCUACUUAAGUGCUCAGGCAGAAUUUUCACAAAUUCCUCGUAAAA ...-------------.....((((..(((((.......))))).........(((..(((((((((((((........)))))))))))))..)))................)))).. ( -23.70) >DroSim_CAF1 29770 106 - 1 CUU-------------AAACAUUAGCAUCAACUUUCGGUGUUGGUUUUCCGUUUUCCCCUUGGGCACUUAAACACCUACUUAAGUGCUCAGGCAGAAUUUUCACAAAUUCCUCGUAAAA ...-------------..(((((((((((.......)))))))))........(((..(((((((((((((........)))))))))))))..)))................)).... ( -25.40) >DroEre_CAF1 35433 106 - 1 CUU-------------AAACAUCAACAUCAACUUUUGGUGUUGGCUCUCCCUUUUCCCCUUCGGCACUUAAACACCUACUUAAGUGCUCAGGCAGAAUUUUGACAAAUUCCUCGUAAAA ...-------------.....(((((((((.....))))))))).............(((..(((((((((........))))))))).)))..((((((....))))))......... ( -23.10) >DroYak_CAF1 34061 106 - 1 CUU-------------AAACAUCAACAUCAACUUUCGGUGUUGGCCCACCGUUUUCCCCUUGGGCACUUAAACACCUACUUAAGUGCUCAGGCAGAAUUUUGACAAACUCCUCGUAAAA ...-------------..((.((((((((.......))))))))......((((((..(((((((((((((........)))))))))))))..)))....))).........)).... ( -27.10) >DroAna_CAF1 39764 112 - 1 CUGAACACCGACGCCGAAACAUUAGCAUUAACUUUUGGUGUUGG-------UUUUCCCCUCGGGCACUUAAACACCUACUUAAGUGCUUAAGCAAAAUUUUCGCACAUUCUCUGUAAAA ..(((.(((((((((((((.............))))))))))))-------).)))..((..(((((((((........)))))))))..))........................... ( -31.12) >consensus CUU_____________AAACAUUAACAUCAACUUUCGGUGUUGGCUCUCCGUUUUCCCCUUGGGCACUUAAACACCUACUUAAGUGCUCAGGCAGAAUUUUCACAAAUUCCUCGUAAAA ..................((.((((((((.......)))))))).........(((..(((((((((((((........)))))))))))))..)))................)).... (-21.82 = -21.78 + -0.04)

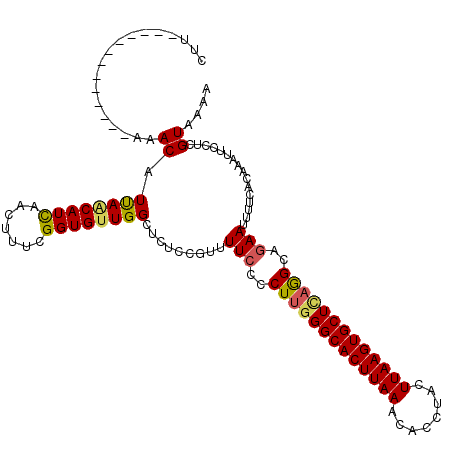
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:26 2006