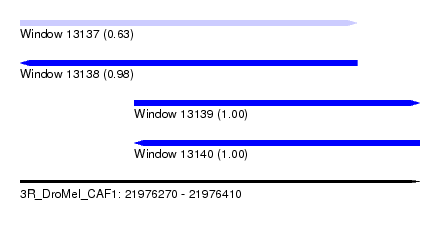
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,976,270 – 21,976,410 |
| Length | 140 |
| Max. P | 0.999779 |

| Location | 21,976,270 – 21,976,388 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 91.86 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -28.74 |
| Energy contribution | -28.98 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21976270 118 + 27905053 UGUCCGUCACGUAUUGAAAUAUAUGACCGGAGACAAGGCCUCGGAAUUACAGCUGCUGGCCAUCAAGUGGGCCAGAUGCCAGAAGUCCAUCUGGCACAUUGCCAUUAUUAAUGACGCA ....((((((((((......)))))...(....)..(((................((((((........)))))).(((((((......)))))))....)))........))))).. ( -35.80) >DroSec_CAF1 15751 118 + 1 UGUCCGUCACGUAUUGAAAUAUAUGACCGGAGACAAGGUCUCGGAAUUACAGCUGCUGGCCAUCAAGUGGGCCAGAUGCCAGAAGUCCAUCUGGCACAUUGCCAUUAUUAAUGACGCA ..(((((((..(((....)))..))))..(((((...))))))))......((..((((((........)))))).(((((((......)))))))(((((.......)))))..)). ( -38.20) >DroEre_CAF1 15640 118 + 1 UGUCCGUCACGUAUUGAAAUAUAUGACCGGAGACAAGGCCUUGGAAUUACAGCUGCUGGCCAUCAAGUGGGCCAGAUGCCAGAAGUCCAUCUGGCACAUUGCCAUUAUUAAUGACGCA ....((((((((((......)))))...(....)..(((..((............((((((........)))))).(((((((......)))))))))..)))........))))).. ( -37.90) >DroYak_CAF1 16479 118 + 1 UGUCCGUCACGUAUUGAAAUAUAUGACCGGAGACAAGGCCUCGGAAUUACAGCUGCUGGCCAUCAAGUGGGCCAGAUGCCAGAAGUCCAUCUGGCACAUUGCCAUUAUUAAUGACGCA ....((((((((((......)))))...(....)..(((................((((((........)))))).(((((((......)))))))....)))........))))).. ( -35.80) >DroAna_CAF1 23242 117 + 1 UGUCCGGCACGUAUUGUAAUAUAUGAGCGGACACGAGGCCUCGGAAUGCCAGAUGCUGGCCAUCAAGUGGAACUGG-GCCAUCUGGGUAUCUGGCGCACUGCCAUUAUUAAUGAUGCA ((((((.(.(((((......))))).)))))))...(((...((..(((((((((((((((....((.....)).)-))).....)))))))))))..)))))............... ( -40.80) >consensus UGUCCGUCACGUAUUGAAAUAUAUGACCGGAGACAAGGCCUCGGAAUUACAGCUGCUGGCCAUCAAGUGGGCCAGAUGCCAGAAGUCCAUCUGGCACAUUGCCAUUAUUAAUGACGCA ....((((((((((......)))))...((......((((.(((........)))..))))....((((.((((((((.........)))))))).)))).))........))))).. (-28.74 = -28.98 + 0.24)


| Location | 21,976,270 – 21,976,388 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 91.86 |
| Mean single sequence MFE | -38.28 |
| Consensus MFE | -32.36 |
| Energy contribution | -33.68 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21976270 118 - 27905053 UGCGUCAUUAAUAAUGGCAAUGUGCCAGAUGGACUUCUGGCAUCUGGCCCACUUGAUGGCCAGCAGCUGUAAUUCCGAGGCCUUGUCUCCGGUCAUAUAUUUCAAUACGUGACGGACA (((((((((((....(((...((((((((......))))))))...)))...))))))))..)))........((((((((...)))))..((((..(((....)))..))))))).. ( -40.20) >DroSec_CAF1 15751 118 - 1 UGCGUCAUUAAUAAUGGCAAUGUGCCAGAUGGACUUCUGGCAUCUGGCCCACUUGAUGGCCAGCAGCUGUAAUUCCGAGACCUUGUCUCCGGUCAUAUAUUUCAAUACGUGACGGACA (((((((((((....(((...((((((((......))))))))...)))...))))))))..)))........((((((((...)))))..((((..(((....)))..))))))).. ( -40.80) >DroEre_CAF1 15640 118 - 1 UGCGUCAUUAAUAAUGGCAAUGUGCCAGAUGGACUUCUGGCAUCUGGCCCACUUGAUGGCCAGCAGCUGUAAUUCCAAGGCCUUGUCUCCGGUCAUAUAUUUCAAUACGUGACGGACA ..((((((.....(((((...((((((((......))))))))((((((........))))))..)))))........((((........))))..............)))))).... ( -38.90) >DroYak_CAF1 16479 118 - 1 UGCGUCAUUAAUAAUGGCAAUGUGCCAGAUGGACUUCUGGCAUCUGGCCCACUUGAUGGCCAGCAGCUGUAAUUCCGAGGCCUUGUCUCCGGUCAUAUAUUUCAAUACGUGACGGACA (((((((((((....(((...((((((((......))))))))...)))...))))))))..)))........((((((((...)))))..((((..(((....)))..))))))).. ( -40.20) >DroAna_CAF1 23242 117 - 1 UGCAUCAUUAAUAAUGGCAGUGCGCCAGAUACCCAGAUGGC-CCAGUUCCACUUGAUGGCCAGCAUCUGGCAUUCCGAGGCCUCGUGUCCGCUCAUAUAUUACAAUACGUGCCGGACA ...............(((..((.(((((((.......((((-((((......)))..)))))..)))))))....))..)))...((((((..((..(((....)))..)).)))))) ( -31.30) >consensus UGCGUCAUUAAUAAUGGCAAUGUGCCAGAUGGACUUCUGGCAUCUGGCCCACUUGAUGGCCAGCAGCUGUAAUUCCGAGGCCUUGUCUCCGGUCAUAUAUUUCAAUACGUGACGGACA (((((((((((....(((...((((((((......))))))))...)))...))))))))..)))........((((((((...)))))..((((..(((....)))..))))))).. (-32.36 = -33.68 + 1.32)


| Location | 21,976,310 – 21,976,410 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 90.80 |
| Mean single sequence MFE | -37.22 |
| Consensus MFE | -30.48 |
| Energy contribution | -31.60 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.998788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21976310 100 + 27905053 UCGGAAUUACAGCUGCUGGCCAUCAAGUGGGCCAGAUGCCAGAAGUCCAUCUGGCACAUUGCCAUUAUUAAUGACGCAAUUGACAUGGCCAGCAUGCAGU ...........((((((((((((......(((.(..(((((((......)))))))..).)))....(((((......))))).)))))))))).))... ( -38.70) >DroSec_CAF1 15791 100 + 1 UCGGAAUUACAGCUGCUGGCCAUCAAGUGGGCCAGAUGCCAGAAGUCCAUCUGGCACAUUGCCAUUAUUAAUGACGCAAUUGACAUGGCCAGCAUGCAGU ...........((((((((((((......(((.(..(((((((......)))))))..).)))....(((((......))))).)))))))))).))... ( -38.70) >DroEre_CAF1 15680 100 + 1 UUGGAAUUACAGCUGCUGGCCAUCAAGUGGGCCAGAUGCCAGAAGUCCAUCUGGCACAUUGCCAUUAUUAAUGACGCAAUUGACAUGGCCAGCAUGCAGG ...........((((((((((((......(((.(..(((((((......)))))))..).)))....(((((......))))).)))))))))).))... ( -38.70) >DroYak_CAF1 16519 100 + 1 UCGGAAUUACAGCUGCUGGCCAUCAAGUGGGCCAGAUGCCAGAAGUCCAUCUGGCACAUUGCCAUUAUUAAUGACGCAAUUGACAUGGCCAGCAUGCAGU ...........((((((((((((......(((.(..(((((((......)))))))..).)))....(((((......))))).)))))))))).))... ( -38.70) >DroAna_CAF1 23282 99 + 1 UCGGAAUGCCAGAUGCUGGCCAUCAAGUGGAACUGG-GCCAUCUGGGUAUCUGGCGCACUGCCAUUAUUAAUGAUGCAAUUGACAUGCCCUGCAUGGACU .(((..(((((((((((((((....((.....)).)-))).....)))))))))))..))).(((((....))))).......(((((...))))).... ( -31.30) >consensus UCGGAAUUACAGCUGCUGGCCAUCAAGUGGGCCAGAUGCCAGAAGUCCAUCUGGCACAUUGCCAUUAUUAAUGACGCAAUUGACAUGGCCAGCAUGCAGU ...........((((((((((((((((((.((((((((.........)))))))).)))((((((.....)))..))).))))..))))))))).))... (-30.48 = -31.60 + 1.12)



| Location | 21,976,310 – 21,976,410 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 90.80 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -32.36 |
| Energy contribution | -33.28 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.85 |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21976310 100 - 27905053 ACUGCAUGCUGGCCAUGUCAAUUGCGUCAUUAAUAAUGGCAAUGUGCCAGAUGGACUUCUGGCAUCUGGCCCACUUGAUGGCCAGCAGCUGUAAUUCCGA ((.((.(((((((((..((((....(((((.....)))))..((.((((((((..(....).)))))))).)).))))))))))))))).))........ ( -40.60) >DroSec_CAF1 15791 100 - 1 ACUGCAUGCUGGCCAUGUCAAUUGCGUCAUUAAUAAUGGCAAUGUGCCAGAUGGACUUCUGGCAUCUGGCCCACUUGAUGGCCAGCAGCUGUAAUUCCGA ((.((.(((((((((..((((....(((((.....)))))..((.((((((((..(....).)))))))).)).))))))))))))))).))........ ( -40.60) >DroEre_CAF1 15680 100 - 1 CCUGCAUGCUGGCCAUGUCAAUUGCGUCAUUAAUAAUGGCAAUGUGCCAGAUGGACUUCUGGCAUCUGGCCCACUUGAUGGCCAGCAGCUGUAAUUCCAA ...((.(((((((((..((((....(((((.....)))))..((.((((((((..(....).)))))))).)).)))))))))))))))........... ( -39.80) >DroYak_CAF1 16519 100 - 1 ACUGCAUGCUGGCCAUGUCAAUUGCGUCAUUAAUAAUGGCAAUGUGCCAGAUGGACUUCUGGCAUCUGGCCCACUUGAUGGCCAGCAGCUGUAAUUCCGA ((.((.(((((((((..((((....(((((.....)))))..((.((((((((..(....).)))))))).)).))))))))))))))).))........ ( -40.60) >DroAna_CAF1 23282 99 - 1 AGUCCAUGCAGGGCAUGUCAAUUGCAUCAUUAAUAAUGGCAGUGCGCCAGAUACCCAGAUGGC-CCAGUUCCACUUGAUGGCCAGCAUCUGGCAUUCCGA .((((.....))))((((((..(((.(((((((...(((.(.((.((((..........))))-.)).).))).)))))))...)))..))))))..... ( -28.40) >consensus ACUGCAUGCUGGCCAUGUCAAUUGCGUCAUUAAUAAUGGCAAUGUGCCAGAUGGACUUCUGGCAUCUGGCCCACUUGAUGGCCAGCAGCUGUAAUUCCGA ..(((((((((((((..((((.((.(((((.....)))))...((((((((......))))))))......)).)))))))))))))..))))....... (-32.36 = -33.28 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:14 2006