| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,791,406 – 2,791,535 |
| Length | 129 |
| Max. P | 0.859375 |

| Location | 2,791,406 – 2,791,505 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 85.69 |
| Mean single sequence MFE | -11.95 |
| Consensus MFE | -9.94 |
| Energy contribution | -9.80 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2791406 99 + 27905053 AGUUAUAUCGAGUAAACGAAAAACAAAAACUUUUUUCCACUUGAGUAACUUAAGUGCCAAUUCCAUCCAAAAACAAA-GAAAAAAAUAAGAAGAACCGAG .......(((.((....((((((........))))))((((((((...)))))))).....................-................))))). ( -11.40) >DroVir_CAF1 118563 92 + 1 AGUUAUAUCGAGUAAACGAAAAACAAAAACUUUUUUCCACUUGAGUAACUUAAGUGCCAAUUCCUCCAAAAACAAAA--------AAAAAAAAUACUAAG .(((.....(((.....((((((........))))))((((((((...)))))))).......)))....)))....--------............... ( -10.60) >DroGri_CAF1 114321 96 + 1 AGUUAUAUCGAGUAAACGAAAAACAAAAACUUUUUUCCACUUGAGUAACUUAAGUGCCAAUUCCUCCAAAAAAAAUA-GAA---GAAGAAAAAAACUAAA .(((...(((......)))..)))......(((((((((((((((...))))))))....(((.((...........-)).---))))))))))...... ( -11.80) >DroSec_CAF1 66809 94 + 1 AGUUAUAUCGAGUAAACGAAAAACAAAAACUUUUUUCCACUUGAGUAACUUAAGUGCCAAUUCCAUCAAAAAAAAAA-A-----AAAAAAAAGAACCGAG .......(((.((....((((((........))))))((((((((...)))))))).....................-.-----..........))))). ( -11.40) >DroSim_CAF1 70835 91 + 1 AGUUAUAUCGAGUAAACGAAAAACAAAAACUUUUUUCCACUUGAGUAACUUAAGUGCCAAUUCCAUCCAAAAAAA---------AAAAAAAAGAACCGAG .......(((.((....((((((........))))))((((((((...))))))))...................---------..........))))). ( -11.40) >DroWil_CAF1 85673 99 + 1 AGUUGUAUCGAGUAAACGAAAAACAAAAACUUUUUUCCACUUGAGUAACUUAAGUGCCAAUUCCCCUUAAAAAGAAGCAAA-CUCAAGACAAAAAACAAA ..((((...((((....((((((........))))))((((((((...))))))))........................)-)))...))))........ ( -15.10) >consensus AGUUAUAUCGAGUAAACGAAAAACAAAAACUUUUUUCCACUUGAGUAACUUAAGUGCCAAUUCCACCAAAAAAAAAA_______AAAAAAAAAAACCAAG ((((((.((((((....((((((........)))))).)))))))))))).................................................. ( -9.94 = -9.80 + -0.14)

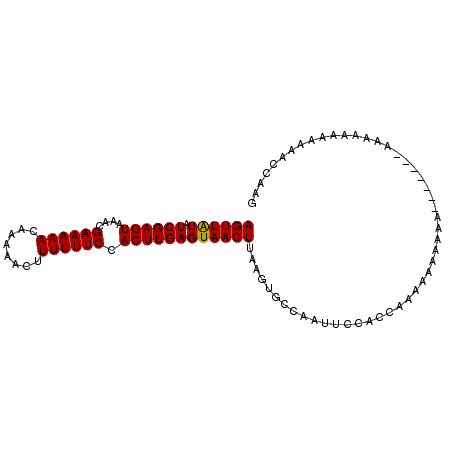
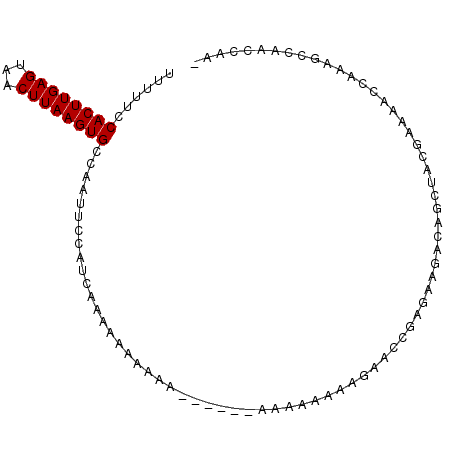
| Location | 2,791,437 – 2,791,535 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 75.56 |
| Mean single sequence MFE | -8.11 |
| Consensus MFE | -6.70 |
| Energy contribution | -6.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2791437 98 + 27905053 UUUUUCCACUUGAGUAACUUAAGUGCCAAUUCCAUCCAAAAACAAAGAAAAAAAUAAGAAGAACCGAGAAGACAGCUACGAAAACCAAAGCCAACCAA- ((((((((((((((...))))))))....(((..............)))................))))))...(((...........))).......- ( -7.74) >DroVir_CAF1 118594 82 + 1 UUUUUCCACUUGAGUAACUUAAGUGCCAAUUCCUCCAAAAACAAAA-------AAAAAAAAUACUAAGAGCACA---AGAAAAACCAACAAA------- ((((((((((((((...)))))))).......(((...........-------..............)))....---.))))))........------- ( -7.81) >DroGri_CAF1 114352 93 + 1 UUUUUCCACUUGAGUAACUUAAGUGCCAAUUCCUCCAAAAAAAAUAGAA---GAAGAAAAAAACUAAAAGCACA---AGAAAAACCAACGCAAACAUGA ((((((((((((((...))))))))....(((.((...........)).---))))))))).............---...................... ( -9.00) >DroSec_CAF1 66840 93 + 1 UUUUUCCACUUGAGUAACUUAAGUGCCAAUUCCAUCAAAAAAAAAAA-----AAAAAAAAGAACCGAGAAGAGAGCUAUGUAAACCAAAGCCAACCAA- ......((((((((...))))))))......................-----....................(.(((.((.....)).))))......- ( -9.40) >DroSim_CAF1 70866 90 + 1 UUUUUCCACUUGAGUAACUUAAGUGCCAAUUCCAUCCAAAAAAA--------AAAAAAAAGAACCGAGAAGACAGCUACGAAAACCAAAGCCAACCAA- .(((((((((((((...))))))))...................--------...............(.((....)).))))))..............- ( -7.10) >DroEre_CAF1 68210 79 + 1 UUUUUCCACUUGAGUAACUUAAGUGCCAAUUCCAUCCAAAAUA---------AAAAAGAAGAACCGAGAAAACAGCUACGCAAACCA----------A- ((((((((((((((...))))))))..................---------.....(......)))))))................----------.- ( -7.60) >consensus UUUUUCCACUUGAGUAACUUAAGUGCCAAUUCCAUCAAAAAAAAAA______AAAAAAAAGAACCGAGAAGACAGCUACGAAAACCAAAGCCAACCAA_ ......((((((((...)))))))).......................................................................... ( -6.70 = -6.70 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:53 2006