| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,967,711 – 21,967,814 |
| Length | 103 |
| Max. P | 0.607123 |

| Location | 21,967,711 – 21,967,814 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.77 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -18.95 |
| Energy contribution | -19.59 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
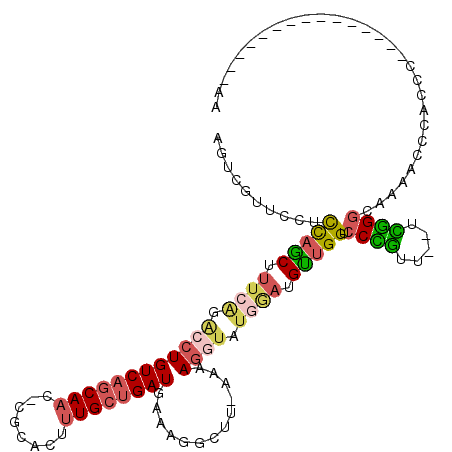
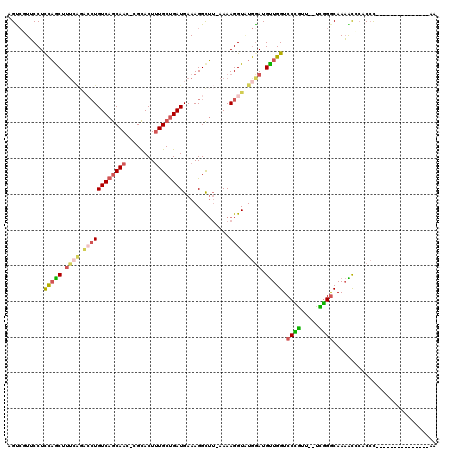
>3R_DroMel_CAF1 21967711 103 - 27905053 AGCCGUUCCUCCAACUUUCAGACCUGUCAGCAAC-CGCACUUUGCUGAUGAAAGGCUU-AAAAGGUAUGGAUGUUGGUCCCGUUUUUCGGACAAAACCCACCC---------------AA .((((..(.(((((((((.((.(((((((((((.-......))))))))...))))).-..))))).)))).).)))).(((.....))).............---------------.. ( -25.20) >DroSec_CAF1 7109 101 - 1 AGUCGUUCCUCCAGCUUUCAGACCUGUCAGCAAC-CGCACUUUGCUGAUGAAAGGCUU-AAAAGGUAUGGAUGUUGGUCCCGUU--UCGGGCAAAACCCACCC---------------AA ..........(((((.((((.((((((((((((.-......)))))))).........-...)))).)))).))))).......--..(((.....)))....---------------.. ( -25.60) >DroSim_CAF1 7112 101 - 1 AGUCGUUCCUCCAGCUUUCAGACCUGUCAGCAAC-CGCACUUUGCUGAUGAAAGGCUU-AAAAGGUAUGGAUGUUGGUCCCGUU--UCGGGCAAAACCCACCC---------------AA ..........(((((.((((.((((((((((((.-......)))))))).........-...)))).)))).))))).......--..(((.....)))....---------------.. ( -25.60) >DroEre_CAF1 7051 99 - 1 UGUCUUUCCUCCAGCUUUCAGACCUGUCAGCAAC-CGCACUUUGCUGAUGAAAGGCUU-AAAAGGUUUGAAUGUUGGUCCUGUU--UCGGGCAAAGC--CCCU---------------CA ..........(((((.(((((((((((((((((.-......)))))))).........-...))))))))).))))).......--..((((...))--))..---------------.. ( -31.00) >DroYak_CAF1 7789 94 - 1 UGUCUUUCCUUCAGCUG-------UGUCAGCAAC-CGCACUUUGCUGAUGAAAGGCUU-AAAAGGUAUGGAUGUUGGUCCUGUU--UUAGGCAAAACACCCCC---------------AA .(((((((..(((((.(-------(((.......-.))))...))))).)))))))..-....((...((.((((...((((..--.))))...)))).))))---------------.. ( -27.40) >DroAna_CAF1 13484 117 - 1 AGC-CUACCUUUCGCUUUGGGGACUGUCAACAAAGGGUACUCUGCCGAUGAAAGGUGAAAAAAGGUGUAGAUGCUGGUCCCAUU--UUGGGCAAUGUACUCACAACAGAAACUGGCAACA .((-(((((((((((((((..(.....)..)))))(((.....)))..))))))))).......(((.((.(((..((((....--..))))...))))))))..........))).... ( -34.30) >consensus AGUCGUUCCUCCAGCUUUCAGACCUGUCAGCAAC_CGCACUUUGCUGAUGAAAGGCUU_AAAAGGUAUGGAUGUUGGUCCCGUU__UCGGGCAAAACCCACCC_______________AA ..........(((((.((((.((((((((((((........)))))))).............)))).)))).))))).((((.....))))............................. (-18.95 = -19.59 + 0.65)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:09 2006