
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,958,441 – 21,958,607 |
| Length | 166 |
| Max. P | 0.878378 |

| Location | 21,958,441 – 21,958,538 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 90.55 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -25.48 |
| Energy contribution | -25.98 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21958441 97 - 27905053 AUUUGUCGCCGACUUAUUAGCGCAGAGGUGCGAACGCUCAAGUAGGUGAAUUAAAGUGCUUCCGCUGUUUACGGUUUGCGAUCUUUGAAUUAAUAUU ....(((((.((((...(((((..((((..(...((((......)))).......)..))))))))).....)))).)))))............... ( -23.40) >DroSec_CAF1 64193 97 - 1 AUUUGUCGCCGACUUAUUAGCGCGGAGGUGCGAACGCUCAAGUAGGUGAAUUAAAGUGCUUCCGCUGUUUACGGUUUGCGAUCUUUAAAUUAAUAUU ....(((((.((((....((((((((((..(...((((......)))).......)..))))))).)))...)))).)))))............... ( -28.70) >DroSim_CAF1 73621 87 - 1 AUUUGUCGCCGACUUAUUAGCGCGGAGGUGCGAACGCUCAAGUAGGUGAAUUAAAGUGCUUCCGCUGUUUACGGUUUGCGAUCU----------AUU ....(((((.((((....((((((((((..(...((((......)))).......)..))))))).)))...)))).)))))..----------... ( -28.70) >DroEre_CAF1 66428 97 - 1 AUUUGUCGCCGACUUUUUAGCGCGGAGGUGCGAAAGAUCAAGUAGGUGAAUUGAAGUGCUUCCGCUGUUUACGGUUUUCGAUCUUUGAUUUAAUAUA ....((((..((((....((((((((((..(...((.(((......))).))...)..))))))).)))...))))..))))............... ( -24.90) >consensus AUUUGUCGCCGACUUAUUAGCGCGGAGGUGCGAACGCUCAAGUAGGUGAAUUAAAGUGCUUCCGCUGUUUACGGUUUGCGAUCUUUGAAUUAAUAUU ....(((((.((((....((((((((((..(......(((......)))......)..))))))).)))...)))).)))))............... (-25.48 = -25.98 + 0.50)



| Location | 21,958,498 – 21,958,607 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.37 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -20.13 |
| Energy contribution | -20.55 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21958498 109 + 27905053 UGAGCGUUCGCACCUCUGCGCUAAUAAGUCGGCGACAAAUCGCUGAUCAUCGACUUGUCCAGCAUGAUUAGGCUUACGAUCU-----UUCGAUCAGCGUGGCGCUAAUCAACAU ..(((((((((.(((....(((.((((((((..((((......)).))..))))))))..)))......))).....((((.-----...)))).))).))))))......... ( -31.40) >DroVir_CAF1 79272 102 + 1 UAUUUGUUCGCAAGCAAUUGCUAAUGGGAUGGUACCAAAUUUUUGAUUAACGCAUG-CUUGCCUUCGAUGGGAUUGCGAGCA-----U------AAACUGGUGCUAAUUCAGUU .........((((....))))...((((.((((((((((((...)))).....(((-(((((..((.....))..)))))))-----)------....)))))))).))))... ( -28.20) >DroSec_CAF1 64250 109 + 1 UGAGCGUUCGCACCUCCGCGCUAAUAAGUCGGCGACAAAUCGCUGAUCAUCGACUUGUCCAGCAUGAUUAGGCUUACGAUCU-----UUCGAUCAGCGUGGUGCUAAUCAACAU .........(((((..((((((.((((((((..((((......)).))..))))))))..)))..............((((.-----...)))).))).))))).......... ( -32.90) >DroSim_CAF1 73668 109 + 1 UGAGCGUUCGCACCUCCGCGCUAAUAAGUCGGCGACAAAUCGCUGAUCAUCGACUUGUCCAGCACGAUUAGGCUUACGAUCU-----UUUGAUCAGCGUGGUGCUAAUCAACAU .........(((((..((((((.((((((((..((((......)).))..))))))))..)))..............((((.-----...)))).))).))))).......... ( -33.50) >DroEre_CAF1 66485 114 + 1 UGAUCUUUCGCACCUCCGCGCUAAAAAGUCGGCGACAAAUCGCUGAUCAUCGACUUGUCCAGUUUGAUUAGGUUUGCGAUCAUCAUCUGCGAUCAGCGUGGUGCUAAUCAACAU .........(((((...(((((.....((((((((....)))))))).........((((((..(((((........)))))....))).))).)))))))))).......... ( -31.60) >DroYak_CAF1 66043 109 + 1 UGAUCUUUCGCACCAACGCGCUAAUAAGUCGGCGACAAAUCGCUGAUCAUCGACUUGUCCAGCAUGAUUAGGCUUACGAUCU-----UUCGAUCAGCGUGGUGCUAAUCAACAU .........((((((.((((((.((((((((..((((......)).))..))))))))..)))..............((((.-----...)))).))))))))).......... ( -35.10) >consensus UGAGCGUUCGCACCUCCGCGCUAAUAAGUCGGCGACAAAUCGCUGAUCAUCGACUUGUCCAGCAUGAUUAGGCUUACGAUCU_____UUCGAUCAGCGUGGUGCUAAUCAACAU .........(((((...(((((.....(((((((......)))))))....((.((((..(((........))).)))))).............)))))))))).......... (-20.13 = -20.55 + 0.42)

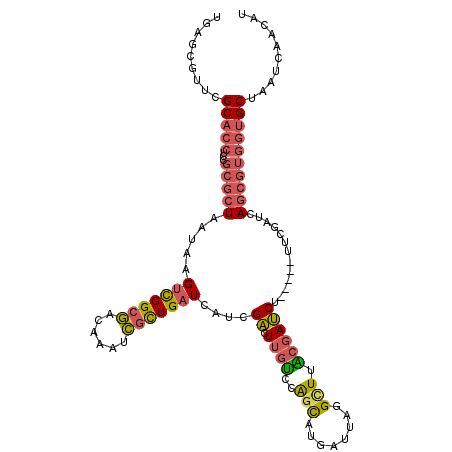

| Location | 21,958,498 – 21,958,607 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.37 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -20.92 |
| Energy contribution | -22.52 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21958498 109 - 27905053 AUGUUGAUUAGCGCCACGCUGAUCGAA-----AGAUCGUAAGCCUAAUCAUGCUGGACAAGUCGAUGAUCAGCGAUUUGUCGCCGACUUAUUAGCGCAGAGGUGCGAACGCUCA ....(((((((.((...((.((((...-----.))))))..)))))))))(((..(((((((((........)))))))))((..(....)..)))))(((((....)).))). ( -36.20) >DroVir_CAF1 79272 102 - 1 AACUGAAUUAGCACCAGUUU------A-----UGCUCGCAAUCCCAUCGAAGGCAAG-CAUGCGUUAAUCAAAAAUUUGGUACCAUCCCAUUAGCAAUUGCUUGCGAACAAAUA (((((.........))))).------.-----((.((((....((......)).(((-((...((((((........((....))....))))))...))))))))).)).... ( -16.40) >DroSec_CAF1 64250 109 - 1 AUGUUGAUUAGCACCACGCUGAUCGAA-----AGAUCGUAAGCCUAAUCAUGCUGGACAAGUCGAUGAUCAGCGAUUUGUCGCCGACUUAUUAGCGCGGAGGUGCGAACGCUCA ..........(((((.(((.((((...-----.))))....((.((((....(.((((((((((........))))))))).).)....)))))))))..)))))......... ( -36.20) >DroSim_CAF1 73668 109 - 1 AUGUUGAUUAGCACCACGCUGAUCAAA-----AGAUCGUAAGCCUAAUCGUGCUGGACAAGUCGAUGAUCAGCGAUUUGUCGCCGACUUAUUAGCGCGGAGGUGCGAACGCUCA ...(((((((((.....))))))))).-----.((.(((..((((..(((((((((..((((((.......((((....)))))))))).)))))))))))))....))).)). ( -39.01) >DroEre_CAF1 66485 114 - 1 AUGUUGAUUAGCACCACGCUGAUCGCAGAUGAUGAUCGCAAACCUAAUCAAACUGGACAAGUCGAUGAUCAGCGAUUUGUCGCCGACUUUUUAGCGCGGAGGUGCGAAAGAUCA ....(((((.(((((.((((((((((.(((....))))).......(((..(((.....))).))))))))))).((((.(((..(....)..))))))))))))....))))) ( -34.90) >DroYak_CAF1 66043 109 - 1 AUGUUGAUUAGCACCACGCUGAUCGAA-----AGAUCGUAAGCCUAAUCAUGCUGGACAAGUCGAUGAUCAGCGAUUUGUCGCCGACUUAUUAGCGCGUUGGUGCGAAAGAUCA ....(((((.(((((((((.((((...-----.))))....((.((((....(.((((((((((........))))))))).).)....))))))))).))))))....))))) ( -39.30) >consensus AUGUUGAUUAGCACCACGCUGAUCGAA_____AGAUCGUAAGCCUAAUCAUGCUGGACAAGUCGAUGAUCAGCGAUUUGUCGCCGACUUAUUAGCGCGGAGGUGCGAACGAUCA ...(((((((((.....))))))))).........((((..((((.....((((((...(((((.(((.(((....)))))).)))))..))))))...))))))))....... (-20.92 = -22.52 + 1.59)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:03 2006