
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,930,738 – 21,930,893 |
| Length | 155 |
| Max. P | 0.966438 |

| Location | 21,930,738 – 21,930,855 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 91.60 |
| Mean single sequence MFE | -34.87 |
| Consensus MFE | -29.17 |
| Energy contribution | -28.73 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.92 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21930738 117 + 27905053 ACACUGAACGUAAAAGUGUUGACAAACAAAUGUGUUCGCCGCACGAUCAUUAACCGCAGAAUUGAAAUACAAUCUGCGGUUUUUGAAGUG--UUUAGCCAGCAAUAUUGUGGUUUAUUU (((((.........)))))....((((.((((((((.((.((((..(((..(((((((((.(((.....))))))))))))..))).)))--)...)).))).)))))...)))).... ( -34.50) >DroSec_CAF1 45326 119 + 1 ACACUGAACAUAAAAGUGUUGACAAAUAAAUCUGUUCACACCACGAUCAUUAACCGCAGAAUUAAAUUACAAUCUGCGGUUUUUGAAGUGGGGUUAGUCAGCAAUAUUAUGGUUUAUUU ........(((((...(((((((.........((....))((((..(((..(((((((((............)))))))))..))).)))).....)))))))...)))))........ ( -36.30) >DroSim_CAF1 54757 119 + 1 ACACUGAACGUAAAAGUGUUGACAAAUAAAUCUAUUCACACCACGAUUAUUAACCGCAGAAUUGAAUUACAAUCUGCGGUUUUUGAAGUGGGGUUAGCCAGCAAUAUUAUGGUUUAUUU (((((.........)))))....(((((((((........((((..(((..(((((((((.(((.....))))))))))))..))).))))((....))...........))))))))) ( -33.80) >consensus ACACUGAACGUAAAAGUGUUGACAAAUAAAUCUGUUCACACCACGAUCAUUAACCGCAGAAUUGAAUUACAAUCUGCGGUUUUUGAAGUGGGGUUAGCCAGCAAUAUUAUGGUUUAUUU ........(((((...(((((.(.((((....))))....((((..(((..(((((((((.(((.....))))))))))))..))).)))).....).)))))...)))))........ (-29.17 = -28.73 + -0.44)

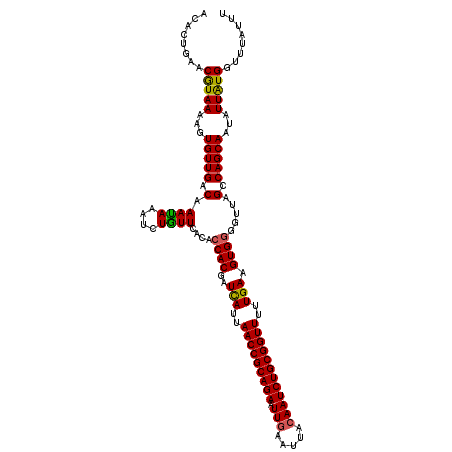

| Location | 21,930,738 – 21,930,855 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.60 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -24.28 |
| Energy contribution | -24.07 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21930738 117 - 27905053 AAAUAAACCACAAUAUUGCUGGCUAAA--CACUUCAAAAACCGCAGAUUGUAUUUCAAUUCUGCGGUUAAUGAUCGUGCGGCGAACACAUUUGUUUGUCAACACUUUUACGUUCAGUGU .................((((((((((--(((.(((..((((((((((((.....))).)))))))))..)))..))).((((((((....))))))))......)))).).))))).. ( -32.20) >DroSec_CAF1 45326 119 - 1 AAAUAAACCAUAAUAUUGCUGACUAACCCCACUUCAAAAACCGCAGAUUGUAAUUUAAUUCUGCGGUUAAUGAUCGUGGUGUGAACAGAUUUAUUUGUCAACACUUUUAUGUUCAGUGU .................(((((.......(((.(((..((((((((((((.....))).)))))))))..)))..)))((((..(((((....)))))..))))........))))).. ( -31.00) >DroSim_CAF1 54757 119 - 1 AAAUAAACCAUAAUAUUGCUGGCUAACCCCACUUCAAAAACCGCAGAUUGUAAUUCAAUUCUGCGGUUAAUAAUCGUGGUGUGAAUAGAUUUAUUUGUCAACACUUUUACGUUCAGUGU .................(((((................((((((((((((.....))).)))))))))......((((((((..(((((....)))))..)))))...))).))))).. ( -27.10) >consensus AAAUAAACCAUAAUAUUGCUGGCUAACCCCACUUCAAAAACCGCAGAUUGUAAUUCAAUUCUGCGGUUAAUGAUCGUGGUGUGAACAGAUUUAUUUGUCAACACUUUUACGUUCAGUGU ...................((((.....((((.(((..((((((((((((.....))).)))))))))..)))..)))).(((((....)))))..))))(((((.........))))) (-24.28 = -24.07 + -0.22)

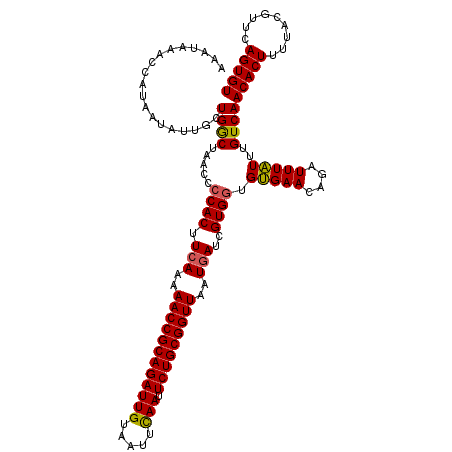
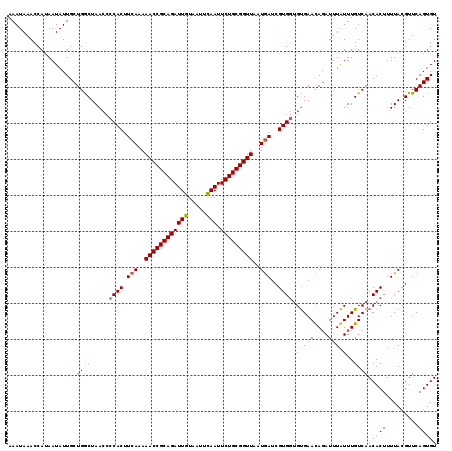
| Location | 21,930,777 – 21,930,893 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 90.37 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -25.29 |
| Energy contribution | -26.30 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21930777 116 + 27905053 CGCACGAUCAUUAACCGCAGAAUUGAAAUACAAUCUGCGGUUUUUGAAGUG--UUUAGCCAGCAAUAUUGUGGUUUAUUUUUAUCACGACAUAGCUAUUUUUGGAGUAAUUAGGCACA .((((..(((..(((((((((.(((.....))))))))))))..))).)))--)...((((((.((.(((((((........))))))).)).))).......((....)).)))... ( -37.40) >DroSec_CAF1 45365 117 + 1 ACCACGAUCAUUAACCGCAGAAUUAAAUUACAAUCUGCGGUUUUUGAAGUGGGGUUAGUCAGCAAUAUUAUGGUUUAUUUUUAACACGAUAAAGCUAUUUUUGGAGUAUGUG-GCACA .((((..(((..(((((((((............)))))))))..))).)))).....((((((..((..((((((((((........))).)))))))...))..))...))-))... ( -30.50) >DroSim_CAF1 54796 117 + 1 ACCACGAUUAUUAACCGCAGAAUUGAAUUACAAUCUGCGGUUUUUGAAGUGGGGUUAGCCAGCAAUAUUAUGGUUUAUUUUUAACACGACAAAGCUAUUUUUGGAGUAAGUA-GCACA .((((..(((..(((((((((.(((.....))))))))))))..))).))))(((.(((((.........))))).)))..............(((((((.......)))))-))... ( -33.50) >consensus ACCACGAUCAUUAACCGCAGAAUUGAAUUACAAUCUGCGGUUUUUGAAGUGGGGUUAGCCAGCAAUAUUAUGGUUUAUUUUUAACACGACAAAGCUAUUUUUGGAGUAAGUA_GCACA .((((..(((..(((((((((.(((.....))))))))))))..))).))))......((((.((((((((.(((((....))).)).))))...)))).)))).............. (-25.29 = -26.30 + 1.01)

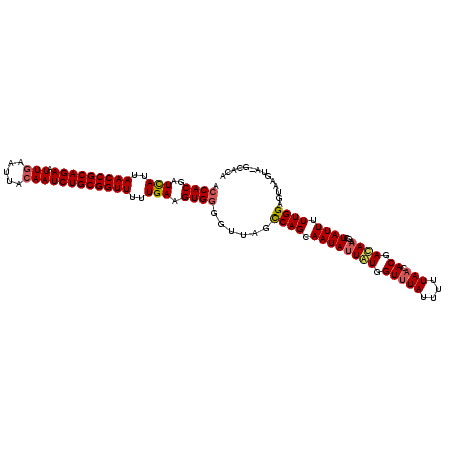

| Location | 21,930,777 – 21,930,893 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 90.37 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -24.63 |
| Energy contribution | -24.63 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21930777 116 - 27905053 UGUGCCUAAUUACUCCAAAAAUAGCUAUGUCGUGAUAAAAAUAAACCACAAUAUUGCUGGCUAAA--CACUUCAAAAACCGCAGAUUGUAUUUCAAUUCUGCGGUUAAUGAUCGUGCG ...(((..(((........)))(((.((((.(((............))).)))).))))))....--(((.(((..((((((((((((.....))).)))))))))..)))..))).. ( -27.60) >DroSec_CAF1 45365 117 - 1 UGUGC-CACAUACUCCAAAAAUAGCUUUAUCGUGUUAAAAAUAAACCAUAAUAUUGCUGACUAACCCCACUUCAAAAACCGCAGAUUGUAAUUUAAUUCUGCGGUUAAUGAUCGUGGU ...((-(((............((((..(((..((............))..)))..))))............(((..((((((((((((.....))).)))))))))..)))..))))) ( -28.60) >DroSim_CAF1 54796 117 - 1 UGUGC-UACUUACUCCAAAAAUAGCUUUGUCGUGUUAAAAAUAAACCAUAAUAUUGCUGGCUAACCCCACUUCAAAAACCGCAGAUUGUAAUUCAAUUCUGCGGUUAAUAAUCGUGGU ...((-(((............(((((..((.((((((...........)))))).)).))))).............((((((((((((.....))).))))))))).......))))) ( -27.90) >consensus UGUGC_UACUUACUCCAAAAAUAGCUUUGUCGUGUUAAAAAUAAACCAUAAUAUUGCUGGCUAACCCCACUUCAAAAACCGCAGAUUGUAAUUCAAUUCUGCGGUUAAUGAUCGUGGU .....................((((..(((.(((............))).)))..)))).......((((.(((..((((((((((((.....))).)))))))))..)))..)))). (-24.63 = -24.63 + 0.01)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:52 2006