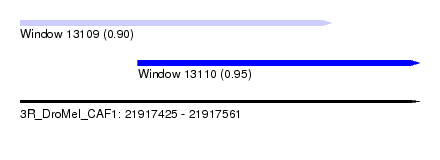
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,917,425 – 21,917,561 |
| Length | 136 |
| Max. P | 0.950368 |

| Location | 21,917,425 – 21,917,531 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.64 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -17.83 |
| Energy contribution | -18.19 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21917425 106 + 27905053 CGCAAAUUUUUAUUGCACUUUUUCCCGGAGCGUCUUGUCUUC-UCCUCUGCUGACU-------UGACUUGUGCGAAUUGUCUGGUUG-CAGACUAGAUAACUCCCUCUGAAAUCC .((((.......)))).........(((((.(..((((((..-...(((((.((((-------.(((...........))).)))))-))))..))))))..).)))))...... ( -26.40) >DroPse_CAF1 35486 97 + 1 CGCAAAUUUUUAUUGCACUU-UUCCUGGAGCGUCUUGUCUU----------UGGCU-------UGACUUGUGCGAAUUGUCUAGUCCCCCGACUAGAUAUCUCUCUCUCACUCCC .((((.......))))....-.....((((.(((..((...----------..)).-------.)))......((..(((((((((....)))))))))..)).......)))). ( -25.40) >DroEre_CAF1 33403 105 + 1 CGCAAAUUUUUAUUGCACUU-UUCCCGGAGCGUCUUGUCUUC-UCCUCCGCUGACU-------UGACUUGUGCGAAUUGUCUGGUCG-CAGACUAGAUAACUCCCUAUGAAAACC .((((.......))))..((-(((..((((.(((..(((..(-......)..))).-------.))).........((((((((((.-..))))))))))))))....))))).. ( -26.00) >DroYak_CAF1 32833 106 + 1 CGCAAAUUUUUAUUGCACUUUUUCCCGGAGCGUCUUGUCUUC-UCCUCGGCUGACU-------UGACUUGUGCGAAUUGUCUGGUCG-CAGACUAGAUAACUCCCUACGAGCACC .((((.......))))..........((.(((((..(((..(-(....))..))).-------.)))(((((.((.((((((((((.-..)))))))))).)).).)))))).)) ( -27.70) >DroAna_CAF1 33087 114 + 1 CGCAAAUUUUUAUUGCACUUUUUCCUGGAGCGUCUUGUCUUCGUCUUCUCCUGACUUGUGUCUUGAGUUGUGCGAAUUGUCUAGUCU-GAGACUACAUAGAUAGCCCUCCAGUUU .((((.......))))........((((((.((..(((((((((...(((..(((....)))..)))....))))).(((.(((((.-..))))).))))))))).))))))... ( -29.70) >DroPer_CAF1 35551 97 + 1 CGCAAAUUUUUAUUGCACUU-UUCCUGGAGCGUCUUGUCUU----------UGGCU-------UGACUUGUGCGAAUUGUCUAGUCCCCCGACUAGAUAUCUCUCUCUCACUCCC .((((.......))))....-.....((((.(((..((...----------..)).-------.)))......((..(((((((((....)))))))))..)).......)))). ( -25.40) >consensus CGCAAAUUUUUAUUGCACUU_UUCCCGGAGCGUCUUGUCUUC_UCCUC_GCUGACU_______UGACUUGUGCGAAUUGUCUAGUCG_CAGACUAGAUAACUCCCUCUCAAAACC .((((.......)))).............(((.(..(((.........................)))..))))((.((((((((((....)))))))))).))............ (-17.83 = -18.19 + 0.36)



| Location | 21,917,465 – 21,917,561 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 70.65 |
| Mean single sequence MFE | -19.32 |
| Consensus MFE | -13.34 |
| Energy contribution | -13.23 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21917465 96 + 27905053 UCUCCUCUGCUGACUUGACUUGUGCGAAUUGUCUGGUUG-CAGACUAGAUAACUCCCUCUGAAAUCCAGCAG--UC-CUCCCCC---CUCUGGUUUUCUC----CUU (((..(((((.((((.(((...........))).)))))-))))..)))...........((((.((((.((--..-.......---)))))).))))..----... ( -21.70) >DroSec_CAF1 32067 97 + 1 UCUCCUCUGCUGACUUGACUUGUGCGAAUUGUCUGGUCG-CAGACUAGAUAACUCCCUCGGAUAUCCAACAG--UACCACUCCC---CCCUGGGUUUCUC----CUU (((..(((((.((((.(((...........))).)))))-))))..)))..........(((.(((((....--..........---...)))))...))----).. ( -22.13) >DroSim_CAF1 41267 100 + 1 UCUCCUCUGCUGACUUGACUUGUGCGAAUUGUCUGGUCG-CAGACUAGAUAACUCCCUCAGAAAUCCAGCAG--UACCACUCCCCCCCCCUGGUUUUCUC----CUU ......((((((..((...(((.(.((.((((((((((.-..)))))))))).)).).))).))..))))))--.((((...........))))......----... ( -22.90) >DroEre_CAF1 33442 87 + 1 UCUCCUCCGCUGACUUGACUUGUGCGAAUUGUCUGGUCG-CAGACUAGAUAACUCCCUAUGAAAACCU-------CCC-----------CCCUUUUUC-CCCCGCUU .......................(((..((((((((((.-..))))))))))........(((((...-------...-----------....)))))-...))).. ( -13.50) >DroYak_CAF1 32873 99 + 1 UCUCCUCGGCUGACUUGACUUGUGCGAAUUGUCUGGUCG-CAGACUAGAUAACUCCCUACGAGCACCC-------CCCACUCCCCACCCUCCAUUUUCUUCUUGAUU .......((......((.((((((.((.((((((((((.-..)))))))))).)).).)))))))...-------.))............................. ( -18.10) >DroPer_CAF1 35590 86 + 1 U---------UGGCUUGACUUGUGCGAAUUGUCUAGUCCCCCGACUAGAUAUCUCUCUCUCACUCCCAUUCGAUUUCUG--------CUCUGCUCUGCUC----GGA .---------(((..(((...(.(.((..(((((((((....)))))))))..)).).))))...)))(((((.....(--------(........))))----))) ( -17.60) >consensus UCUCCUCUGCUGACUUGACUUGUGCGAAUUGUCUGGUCG_CAGACUAGAUAACUCCCUCUGAAAUCCA_CAG__UCCCACUCCC___CCCUGGUUUUCUC____CUU .........................((.((((((((((....)))))))))).)).................................................... (-13.34 = -13.23 + -0.11)

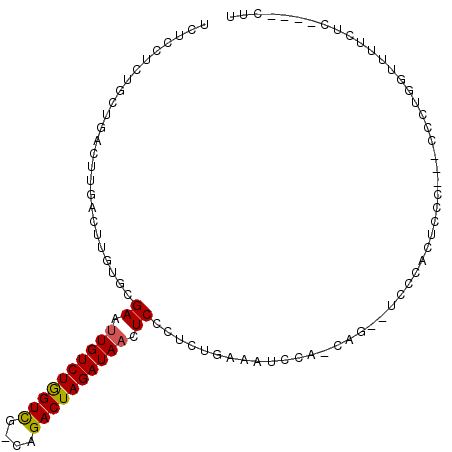
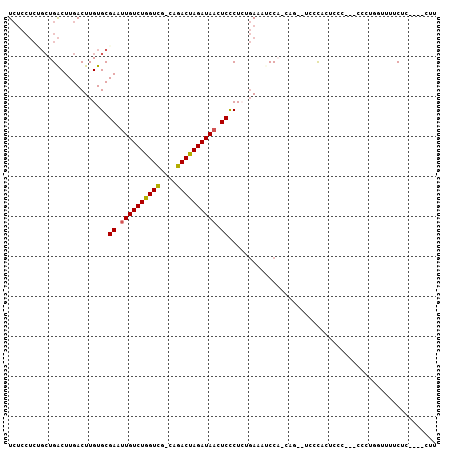
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:46 2006