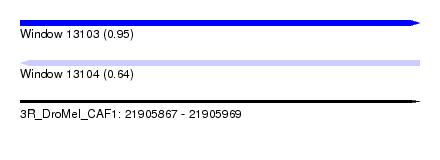
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,905,867 – 21,905,969 |
| Length | 102 |
| Max. P | 0.953550 |

| Location | 21,905,867 – 21,905,969 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -22.03 |
| Consensus MFE | -17.79 |
| Energy contribution | -18.23 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21905867 102 + 27905053 AAAAAAACAAAAAAACUAA---ACCUCCCCAUCCCCCAAA--AAACCCGUCGGAUGACGCAGACGCCACAGUCGUUUGUGGGGUUUGUUUUUCUUCAUUUUUUUUUC (((((((...((((((.((---((((...(((((..(...--......)..))))).(((((((((....).)))))))))))))))))))).....)))))))... ( -23.60) >DroSec_CAF1 20651 90 + 1 ------------AAACAAA---ACCUCCCGAUCCCCCAAA--AAACCCGUCGGAUGACGCAGACGCCACAGUCGUUUGUGGGGUUGGUUUUUCUUAAUUUUUUUUUC ------------.......---.....((((((((.((((--.....((((....))))..(((......))).)))).)))))))).................... ( -22.20) >DroSim_CAF1 20694 90 + 1 ------------AAACAAA---ACCUCCCGAUCCCCCAAA--AAACCCGUCGGAUGACGCAGACGCCACAGUCGUUUGUGGGGUUUGUUUUUUUUAAUUUUUUUUUC ------------.......---.....(((((........--......))))).....((((((.((((((....)))))).))))))................... ( -20.24) >DroEre_CAF1 21888 85 + 1 ------------AAAAAAAACAACCUCCCGAUCCCCCAA---AAACCCGUCGGAUGACGCAGACGCCACAGUCUUUUGCGGGGUUUGCUUUUCUUUCGUU------- ------------......(((......(((((.......---......))))).....((((((.((.(((....))).)).)))))).........)))------- ( -19.22) >DroYak_CAF1 21215 89 + 1 ------------AAAAAAAACAAACUCCCGAUCCCCCAAAAAAAACCCGUCGGAUGACGCAGACGCCACAGUCGUUUGUGGGGUUUGUUUUUCUUCAAUUC------ ------------...(((((((((((((((((................)))))....(((((((((....).)))))))))))))))))))).........------ ( -24.89) >consensus ____________AAACAAA___ACCUCCCGAUCCCCCAAA__AAACCCGUCGGAUGACGCAGACGCCACAGUCGUUUGUGGGGUUUGUUUUUCUUAAUUUUUUUUUC ...........................(((((................))))).....((((((.((((((....)))))).))))))................... (-17.79 = -18.23 + 0.44)

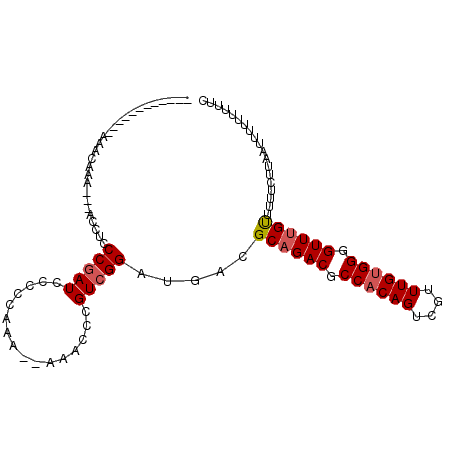
| Location | 21,905,867 – 21,905,969 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -17.50 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21905867 102 - 27905053 GAAAAAAAAAUGAAGAAAAACAAACCCCACAAACGACUGUGGCGUCUGCGUCAUCCGACGGGUUU--UUUGGGGGAUGGGGAGGU---UUAGUUUUUUUGUUUUUUU ...(((((((..((((((...((((((((((......)))))..(((.((((..((((.......--.))))..)))).))))))---))..))))))..))))))) ( -31.30) >DroSec_CAF1 20651 90 - 1 GAAAAAAAAAUUAAGAAAAACCAACCCCACAAACGACUGUGGCGUCUGCGUCAUCCGACGGGUUU--UUUGGGGGAUCGGGAGGU---UUUGUUU------------ ..............((((((((.((.(((((......))))).))...((((....)))))))))--)))..((((((....)))---)))....------------ ( -22.70) >DroSim_CAF1 20694 90 - 1 GAAAAAAAAAUUAAAAAAAACAAACCCCACAAACGACUGUGGCGUCUGCGUCAUCCGACGGGUUU--UUUGGGGGAUCGGGAGGU---UUUGUUU------------ .................(((((((((((.((((.((((((((((....))))))......)))).--))))))))(((....)))---)))))))------------ ( -24.20) >DroEre_CAF1 21888 85 - 1 -------AACGAAAGAAAAGCAAACCCCGCAAAAGACUGUGGCGUCUGCGUCAUCCGACGGGUUU---UUGGGGGAUCGGGAGGUUGUUUUUUUU------------ -------.....((((((((((..((((.((((((.((((((((....)))).....)))).)))---)))))))(((....)))))))))))))------------ ( -26.90) >DroYak_CAF1 21215 89 - 1 ------GAAUUGAAGAAAAACAAACCCCACAAACGACUGUGGCGUCUGCGUCAUCCGACGGGUUUUUUUUGGGGGAUCGGGAGUUUGUUUUUUUU------------ ------......((((((((((((((((...........(((((....)))))(((..((((.....))))..)))..))).)))))))))))))------------ ( -28.40) >consensus GAAAAAAAAAUGAAGAAAAACAAACCCCACAAACGACUGUGGCGUCUGCGUCAUCCGACGGGUUU__UUUGGGGGAUCGGGAGGU___UUUGUUU____________ ........................((((.((((.((((((((((....))))))......))))...))))))))................................ (-17.50 = -17.70 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:40 2006