
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,862,066 – 21,862,159 |
| Length | 93 |
| Max. P | 0.658363 |

| Location | 21,862,066 – 21,862,159 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.11 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -16.76 |
| Energy contribution | -16.79 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
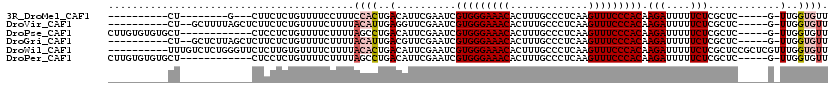
>3R_DroMel_CAF1 21862066 93 + 27905053 AACACCAA-C-----GAGCGAGAAAAAUCUUGUGGGAAACUUGAGGGCAAAGUGUUUCCCACGAUUCGAAUGUCAGUGGAAAGGAAAACAGAGAAG---C--------AG---------- ..(((..(-(-----(..((((.......(((((((((((.............)))))))))))))))..)))..)))..................---.--------..---------- ( -22.53) >DroVir_CAF1 49494 102 + 1 AACACCAA-C-----GAGCGAGAAAAAUCUUGUGGGAAACUUGAGGGCAAAGUGUUUCCCACGAUUCGAACCUCAAUGUAAAAGAAAACAGAGAAGAGCUAAAAGC--AG---------- ........-(-----((((((((....))))(((((((((.............)))))))))).))))...(((..(((........))))))....((.....))--..---------- ( -21.52) >DroPse_CAF1 47479 102 + 1 AACACCAA-C-----GAGCGAGAAAAAUCUUGUGGGAAACUUGAGGGCAAAGUGUUUCCCACGAUUCGAAUGUCAGGCUAAAAGAAAACAGAGGAG------------AGCACACACAAG ........-(-----((((((((....))))(((((((((.............)))))))))).))))..(((...(((.................------------)))..))).... ( -20.05) >DroGri_CAF1 43372 102 + 1 AACACCAA-C-----GAGCGAGAAAAAUCUUGUGGGAAACUUGAGGGCAAAGUGUUUCCCACGAUUCGAACGUCAAUGUAAAAGAAAACAGAGAAGAGCUAAGAGC--AG---------- .(((...(-(-----(..((((.......(((((((((((.............)))))))))))))))..)))...)))..................((.....))--..---------- ( -22.33) >DroWil_CAF1 50797 110 + 1 AACACCAAACGAGCGGAGCGAGAAAAAUCUUGUGGGAAACUUGAGGGCAAAGUGUUUCCCACGAUUCGAAUGUCAGUGUAAAAGAAAACACAAGAGAACCCAGAGACAAA---------- .........((((((..((((((....))))))(((((((.............))))))).)).))))..((((.((((........))))..(......)...))))..---------- ( -26.02) >DroPer_CAF1 47751 102 + 1 AACACCAA-C-----GAGCGAGAAAAAUCUUGUGGGAAACUUGAGGGCAAAGUGUUUCCCACGAUUCGAAUGUCAGGCUAAAAGAAAACAGAGGAG------------AGCACACACAAG ........-(-----((((((((....))))(((((((((.............)))))))))).))))..(((...(((.................------------)))..))).... ( -20.05) >consensus AACACCAA_C_____GAGCGAGAAAAAUCUUGUGGGAAACUUGAGGGCAAAGUGUUUCCCACGAUUCGAAUGUCAGUGUAAAAGAAAACAGAGAAG____________AG__________ ...........................(((((((((((((.............)))))))))(((......))).......))))................................... (-16.76 = -16.79 + 0.03)



| Location | 21,862,066 – 21,862,159 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.11 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -15.54 |
| Energy contribution | -16.15 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21862066 93 - 27905053 ----------CU--------G---CUUCUCUGUUUUCCUUUCCACUGACAUUCGAAUCGUGGGAAACACUUUGCCCUCAAGUUUCCCACAAGAUUUUUCUCGCUC-----G-UUGGUGUU ----------..--------.---..................(((..((...(((...(((((((((.............)))))))))..((....)))))...-----)-)..))).. ( -22.82) >DroVir_CAF1 49494 102 - 1 ----------CU--GCUUUUAGCUCUUCUCUGUUUUCUUUUACAUUGAGGUUCGAAUCGUGGGAAACACUUUGCCCUCAAGUUUCCCACAAGAUUUUUCUCGCUC-----G-UUGGUGUU ----------..--((.....))..................((((..(.(..(((...(((((((((.............)))))))))..((....)))))..)-----.-)..)))). ( -23.72) >DroPse_CAF1 47479 102 - 1 CUUGUGUGUGCU------------CUCCUCUGUUUUCUUUUAGCCUGACAUUCGAAUCGUGGGAAACACUUUGCCCUCAAGUUUCCCACAAGAUUUUUCUCGCUC-----G-UUGGUGUU ............------------.................((((..((...(((...(((((((((.............)))))))))..((....)))))...-----)-)..).))) ( -19.92) >DroGri_CAF1 43372 102 - 1 ----------CU--GCUCUUAGCUCUUCUCUGUUUUCUUUUACAUUGACGUUCGAAUCGUGGGAAACACUUUGCCCUCAAGUUUCCCACAAGAUUUUUCUCGCUC-----G-UUGGUGUU ----------..--((.....))..................((((..(((..(((...(((((((((.............)))))))))..((....)))))..)-----)-)..)))). ( -27.32) >DroWil_CAF1 50797 110 - 1 ----------UUUGUCUCUGGGUUCUCUUGUGUUUUCUUUUACACUGACAUUCGAAUCGUGGGAAACACUUUGCCCUCAAGUUUCCCACAAGAUUUUUCUCGCUCCGCUCGUUUGGUGUU ----------...((((...(((((((..((((........)))).)).....)))))(((((((((.............))))))))).)))).......((.(((......))).)). ( -23.52) >DroPer_CAF1 47751 102 - 1 CUUGUGUGUGCU------------CUCCUCUGUUUUCUUUUAGCCUGACAUUCGAAUCGUGGGAAACACUUUGCCCUCAAGUUUCCCACAAGAUUUUUCUCGCUC-----G-UUGGUGUU ............------------.................((((..((...(((...(((((((((.............)))))))))..((....)))))...-----)-)..).))) ( -19.92) >consensus __________CU____________CUCCUCUGUUUUCUUUUACACUGACAUUCGAAUCGUGGGAAACACUUUGCCCUCAAGUUUCCCACAAGAUUUUUCUCGCUC_____G_UUGGUGUU .........................................((((..(..........(((((((((.............))))))))).(((....)))............)..)))). (-15.54 = -16.15 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:30 2006