
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,859,026 – 21,859,158 |
| Length | 132 |
| Max. P | 0.993231 |

| Location | 21,859,026 – 21,859,128 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -18.00 |
| Consensus MFE | -16.15 |
| Energy contribution | -16.27 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
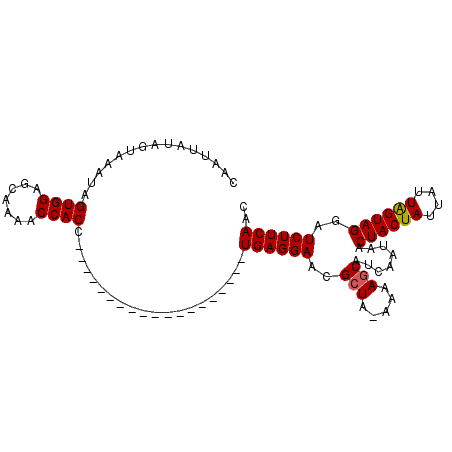
>3R_DroMel_CAF1 21859026 102 + 27905053 CAAUUAUAGUAAAUAGUGGAGCAAAACCACCUUCAAUGUUAGUUGCAACAUGAGGAACGCUA-CAAAACAUCAACAACUACUAUUAUUAGUAGCAUCUUCAAC ......((((.....((((.......))))(((((.((((......)))))))))...))))-..............((((((....)))))).......... ( -17.30) >DroSec_CAF1 42572 83 + 1 CAAUUAUAGUAAAUAGUGGAGAAAAACCACC-------------------UGAGGAACGCUA-AAAAGCAUCAAUAACUACUAUUAUUAGUAGGAUCUUCAAC ...............((((.......)))).-------------------((((((.((((.-...)))........((((((....))))))).)))))).. ( -18.50) >DroSim_CAF1 35686 84 + 1 CAAUUAUAGUAAAUAGUGGAGCAAAACCACC-------------------UGAGGAACGCUACAAAAGCAUCAAUAACUACUAUUAUUGGUAGGAUCUUCAAC ...............((((.......)))).-------------------((((((.((((.....)))........((((((....))))))).)))))).. ( -18.20) >consensus CAAUUAUAGUAAAUAGUGGAGCAAAACCACC___________________UGAGGAACGCUA_AAAAGCAUCAAUAACUACUAUUAUUAGUAGGAUCUUCAAC ...............((((.......))))....................((((((..(((.....)))........((((((....))))))..)))))).. (-16.15 = -16.27 + 0.11)

| Location | 21,859,026 – 21,859,128 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 82.01 |
| Mean single sequence MFE | -19.30 |
| Consensus MFE | -15.64 |
| Energy contribution | -15.20 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21859026 102 - 27905053 GUUGAAGAUGCUACUAAUAAUAGUAGUUGUUGAUGUUUUG-UAGCGUUCCUCAUGUUGCAACUAACAUUGAAGGUGGUUUUGCUCCACUAUUUACUAUAAUUG .........(((((((....)))))))..(..((((((((-((((((.....)))))))))..)))))..).(((((.......))))).............. ( -24.60) >DroSec_CAF1 42572 83 - 1 GUUGAAGAUCCUACUAAUAAUAGUAGUUAUUGAUGCUUUU-UAGCGUUCCUCA-------------------GGUGGUUUUUCUCCACUAUUUACUAUAAUUG ..(((.((..((((((....))))))......(((((...-.))))))).)))-------------------(((((.......))))).............. ( -15.90) >DroSim_CAF1 35686 84 - 1 GUUGAAGAUCCUACCAAUAAUAGUAGUUAUUGAUGCUUUUGUAGCGUUCCUCA-------------------GGUGGUUUUGCUCCACUAUUUACUAUAAUUG ..............((((.(((((((.....((((((.....)))))).....-------------------(((((.......)))))..))))))).)))) ( -17.40) >consensus GUUGAAGAUCCUACUAAUAAUAGUAGUUAUUGAUGCUUUU_UAGCGUUCCUCA___________________GGUGGUUUUGCUCCACUAUUUACUAUAAUUG ..............((((.(((((((.....((((((.....))))))........................(((((.......)))))..))))))).)))) (-15.64 = -15.20 + -0.44)



| Location | 21,859,066 – 21,859,158 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.28 |
| Mean single sequence MFE | -17.27 |
| Consensus MFE | -5.64 |
| Energy contribution | -5.48 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21859066 92 + 27905053 AGUUGCAACAUGAGGAA---CGCUA-CAAA--------------ACAUCAACAACUACUAUUAUUAGUAGCAUCUUCAACGUCGAUAACUAUA----------CAGCAAUACAAUAAGGA .(((((....((((((.---.((((-(...--------------......................))))).))))))..((.....))....----------..))))).......... ( -14.11) >DroSec_CAF1 42603 77 + 1 ----------UGAGGAA---CGCUA-AAAA--------------GCAUCAAUAACUACUAUUAUUAGUAGGAUCUUCAACGUCGAUAACUAUA----------CAGCA-----AUAAGGA ----------((((((.---((((.-...)--------------))........((((((....))))))).))))))...............----------.....-----....... ( -14.60) >DroSim_CAF1 35717 83 + 1 ----------UGAGGAA---CGCUACAAAA--------------GCAUCAAUAACUACUAUUAUUGGUAGGAUCUUCAACGUCGAUAACUAUA----------CAGCAAUACAAUAAGGA ----------((((((.---((((.....)--------------))........((((((....))))))).))))))...............----------................. ( -14.30) >DroEre_CAF1 36277 113 + 1 AGUUGCAACAGGAGUAA---CACUA-CAAAAGAUGAAGCGGCAAACGUCAAUAACUACUA---UUAGUAGCAUCUUCAACAUCAAUAACUAUAACAUUAAAUCCAACAAUACAAUUGGGA .(((((.......))))---)....-...((((((....(((....))).....((((..---...)))))))))).........................(((((........))))). ( -16.70) >DroYak_CAF1 40164 119 + 1 AGUUGCAACAGGAGGAGCUGCACUA-CAAAAGACGAAGCAGCUAACCUCAAUGACUACUAUUAUUAGUAGCAUCUUCAACAUCAAAAACUAUAACAUUAAAUCCAGCAAUAUAACAAGGA .(((((.....((((((((((....-...........))))))..)))).(((.((((((....)))))))))................................))))).......... ( -26.66) >consensus AGUUGCAACAUGAGGAA___CGCUA_CAAA______________ACAUCAAUAACUACUAUUAUUAGUAGCAUCUUCAACGUCGAUAACUAUA__________CAGCAAUACAAUAAGGA ......................................................((((((....)))))).................................................. ( -5.64 = -5.48 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:28 2006