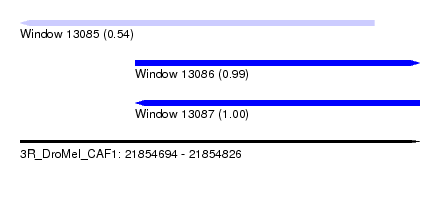
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,854,694 – 21,854,826 |
| Length | 132 |
| Max. P | 0.999948 |

| Location | 21,854,694 – 21,854,811 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.63 |
| Mean single sequence MFE | -22.72 |
| Consensus MFE | -12.07 |
| Energy contribution | -13.15 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
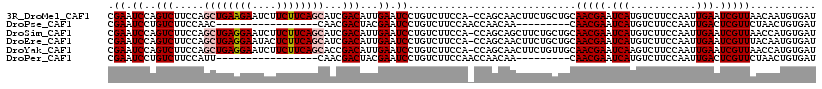
>3R_DroMel_CAF1 21854694 117 - 27905053 CGAAUCCAGUCUUCCAGCUGAAGAAUCUUCUUCAGCAUCGACAUUGAAUCCUGUCUUCCA-CCAGCAACUUCUGCUGCAACGAAUCAUGUCUUCCAAUUGAAUCGUUAACAAUGUGAU ................((((((((....))))))))(((.((((((..............-.(((((.....))))).(((((.(((...........))).)))))..))))))))) ( -30.00) >DroPse_CAF1 39114 92 - 1 CGAAUCCUGUCUUCCAAC-----------------CAACGACUACGAAUCCUGUCUUCCAACCAACAA---------CAACGAAUCAUGUCUUCCAAUUGACUCGUUCUAACUGUGAU .((.((..(((.......-----------------....)))...)).))..................---------.(((((.(((...........))).)))))........... ( -7.10) >DroSim_CAF1 31388 117 - 1 CGAAUCCAGUCUUCCAGCUGAGGAAUCUUCUUCAGCAUCGACAUUGAAUCCUGUCUUCCA-CCAGCAGCUUCUGCUGCAACGAAUCAUGUCUUCCAAUUGAAUCGUUAACCAUGUGAU .(((..(((.......((((((((....)))))))).(((....)))...)))..)))((-(..(((((....)))))(((((.(((...........))).)))))......))).. ( -32.00) >DroEre_CAF1 31907 117 - 1 CGAAUCCAGUCUUCCAGCUGAGGAAUACUCUUCAGCAUCGACAUUGAAUCCUGUCUUCCA-CCAGCAACUUCUGCUGCAACGAAUCAUGUCUUCCAAUUGAAUCGUUUACAAUGUGAU ................((((((((....))))))))(((.((((((..............-.(((((.....))))).(((((.(((...........))).)))))..))))))))) ( -29.30) >DroYak_CAF1 35701 117 - 1 CGAAUCCAGUCUUCCAGCUGAGGAAUCUUCUUCAGCACCGACAUUGAAUCCUGUCUUCCA-CCAGCAACUUCUGUUGCAACGAAUCAAGUCUUCCAAUUGAAUCGUUAACCAUGUGAU .(((..(((..(((..((((((((....)))))))).........)))..)))..)))((-(..(((((....)))))(((((.((((.........)))).)))))......))).. ( -30.50) >DroPer_CAF1 39495 92 - 1 CGAAUCCUGUCUUCCAUU-----------------CAACGACUACGAAUCCUGUCUUCCAACCAACAA---------CAACGAAUCAUGUCUUCCAAUUGACUCGUUCUAACUGUGAU .................(-----------------((..(((..........))).............---------.(((((.(((...........))).))))).......))). ( -7.40) >consensus CGAAUCCAGUCUUCCAGCUGAGGAAUCUUCUUCAGCAUCGACAUUGAAUCCUGUCUUCCA_CCAGCAACUUCUGCUGCAACGAAUCAUGUCUUCCAAUUGAAUCGUUAACAAUGUGAU .((.((..(((.....((((((((....))))))))...)))...)).))............................(((((.(((...........))).)))))........... (-12.07 = -13.15 + 1.08)

| Location | 21,854,732 – 21,854,826 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 96.81 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -26.30 |
| Energy contribution | -25.98 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21854732 94 + 27905053 UUGCAGCAGAAGUUGCUGGUGGAAGACAGGAUUCAAUGUCGAUGCUGAAGAAGAUUCUUCAGCUGGAAGACUGGAUUCGAUGUCGAUGAUGAUG ..(((((....)))))........(((((((((((..(((...((((((((....)))))))).....))))))))))..)))).......... ( -29.70) >DroSec_CAF1 38319 94 + 1 UUGCAGCAGAAGUUGCUGGUGGAAGACAGGAUUCAAUGUCGAUGCUGAAGAAGAUUCCUCAGCUGGAAGACUGGAUUCGAUGUCGAUGAUGAUG ..(((((....)))))........(((((((((((..(((...(((((.((....)).))))).....))))))))))..)))).......... ( -25.60) >DroSim_CAF1 31426 94 + 1 UUGCAGCAGAAGCUGCUGGUGGAAGACAGGAUUCAAUGUCGAUGCUGAAGAAGAUUCCUCAGCUGGAAGACUGGAUUCGAUGUCGAUGAUGAUG ..(((((....)))))........(((((((((((..(((...(((((.((....)).))))).....))))))))))..)))).......... ( -28.00) >DroEre_CAF1 31945 94 + 1 UUGCAGCAGAAGUUGCUGGUGGAAGACAGGAUUCAAUGUCGAUGCUGAAGAGUAUUCCUCAGCUGGAAGACUGGAUUCGAUGUCGUUGAUGAUG ..(((((....)))))..((.((.(((((((((((..(((...(((((.(......).))))).....))))))))))..)))).)).)).... ( -25.90) >DroYak_CAF1 35739 94 + 1 UUGCAACAGAAGUUGCUGGUGGAAGACAGGAUUCAAUGUCGGUGCUGAAGAAGAUUCCUCAGCUGGAAGACUGGAUUCGAUGUCGUUGAUGAUG ..(((((....)))))..((.((.(((((((((((..(((...(((((.((....)).))))).....))))))))))..)))).)).)).... ( -26.30) >consensus UUGCAGCAGAAGUUGCUGGUGGAAGACAGGAUUCAAUGUCGAUGCUGAAGAAGAUUCCUCAGCUGGAAGACUGGAUUCGAUGUCGAUGAUGAUG ..(((((....)))))........(((((((((((..(((...(((((.((....)).))))).....))))))))))..)))).......... (-26.30 = -25.98 + -0.32)



| Location | 21,854,732 – 21,854,826 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 96.81 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -22.78 |
| Energy contribution | -22.46 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.97 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.77 |
| SVM RNA-class probability | 0.999948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
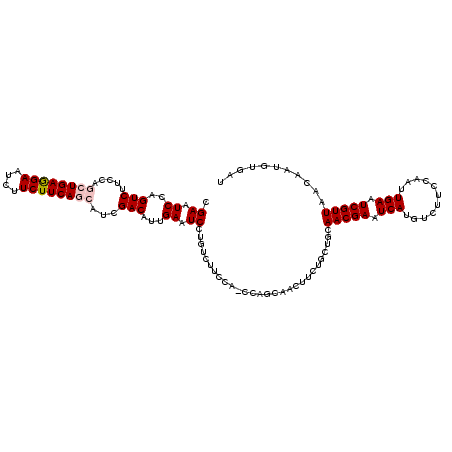
>3R_DroMel_CAF1 21854732 94 - 27905053 CAUCAUCAUCGACAUCGAAUCCAGUCUUCCAGCUGAAGAAUCUUCUUCAGCAUCGACAUUGAAUCCUGUCUUCCACCAGCAACUUCUGCUGCAA ..........((((..((.((..(((.....((((((((....))))))))...)))...)).)).))))......(((((.....)))))... ( -23.00) >DroSec_CAF1 38319 94 - 1 CAUCAUCAUCGACAUCGAAUCCAGUCUUCCAGCUGAGGAAUCUUCUUCAGCAUCGACAUUGAAUCCUGUCUUCCACCAGCAACUUCUGCUGCAA ..........((((..((.((..(((.....((((((((....))))))))...)))...)).)).))))......(((((.....)))))... ( -22.70) >DroSim_CAF1 31426 94 - 1 CAUCAUCAUCGACAUCGAAUCCAGUCUUCCAGCUGAGGAAUCUUCUUCAGCAUCGACAUUGAAUCCUGUCUUCCACCAGCAGCUUCUGCUGCAA ..........((((..((.((..(((.....((((((((....))))))))...)))...)).)).))))........(((((....))))).. ( -26.70) >DroEre_CAF1 31945 94 - 1 CAUCAUCAACGACAUCGAAUCCAGUCUUCCAGCUGAGGAAUACUCUUCAGCAUCGACAUUGAAUCCUGUCUUCCACCAGCAACUUCUGCUGCAA ..........((((..((.((..(((.....((((((((....))))))))...)))...)).)).))))......(((((.....)))))... ( -22.90) >DroYak_CAF1 35739 94 - 1 CAUCAUCAACGACAUCGAAUCCAGUCUUCCAGCUGAGGAAUCUUCUUCAGCACCGACAUUGAAUCCUGUCUUCCACCAGCAACUUCUGUUGCAA ..........((((..((.((..(((.....((((((((....))))))))...)))...)).)).))))........(((((....))))).. ( -24.30) >consensus CAUCAUCAUCGACAUCGAAUCCAGUCUUCCAGCUGAGGAAUCUUCUUCAGCAUCGACAUUGAAUCCUGUCUUCCACCAGCAACUUCUGCUGCAA ..........((((..((.((..(((.....((((((((....))))))))...)))...)).)).))))......(((((.....)))))... (-22.78 = -22.46 + -0.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:24 2006