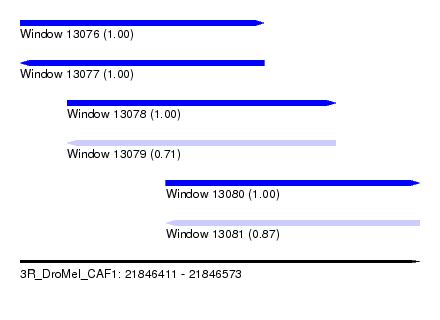
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,846,411 – 21,846,573 |
| Length | 162 |
| Max. P | 0.999990 |

| Location | 21,846,411 – 21,846,510 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 96.41 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -22.73 |
| Energy contribution | -22.90 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.58 |
| SVM RNA-class probability | 0.999990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21846411 99 + 27905053 AACA---------GCAACAACAUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUC ....---------((............))...((..(((((.((...........)).)))))........((((((((((...........)))))))))))).... ( -23.30) >DroEre_CAF1 23320 99 + 1 AACA---------GCAACAACAUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUC ....---------((............))...((..(((((.((...........)).)))))........((((((((((...........)))))))))))).... ( -23.30) >DroWil_CAF1 30479 108 + 1 AACACCAACAACAUCAACAACAUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUC ...(((..............................(((((.((...........)).)))))........((((((((((...........)))))))))))))... ( -24.60) >DroYak_CAF1 26912 99 + 1 AACA---------GCAACAACAUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUC ....---------((............))...((..(((((.((...........)).)))))........((((((((((...........)))))))))))).... ( -23.30) >DroAna_CAF1 27493 99 + 1 AACA---------GCAACAACAUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUC ....---------((............))...((..(((((.((...........)).)))))........((((((((((...........)))))))))))).... ( -23.30) >DroPer_CAF1 29036 98 + 1 AACA---------GCAACAACAUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGA-ACACACGACAAAUCUGUUUCCCACGGUCUC ....---------((............))...((..(((((.((...........)).)))))........((((((-(.(((.((....)))))))))))))).... ( -21.10) >consensus AACA_________GCAACAACAUUUCAGCAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUC ................................((..(((((.((...........)).)))))........((((((((((...........)))))))))))).... (-22.73 = -22.90 + 0.17)

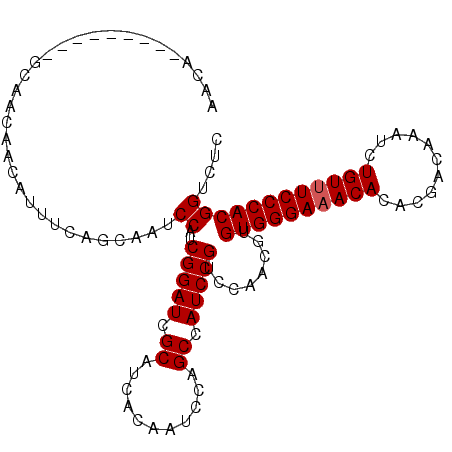

| Location | 21,846,411 – 21,846,510 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 96.41 |
| Mean single sequence MFE | -33.63 |
| Consensus MFE | -33.33 |
| Energy contribution | -33.50 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.29 |
| SVM RNA-class probability | 0.999861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21846411 99 - 27905053 GAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAUGUUGUUGC---------UGUU ......((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))(((.((......)).)))..---------.... ( -33.80) >DroEre_CAF1 23320 99 - 1 GAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAUGUUGUUGC---------UGUU ......((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))(((.((......)).)))..---------.... ( -33.80) >DroWil_CAF1 30479 108 - 1 GAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAUGUUGUUGAUGUUGUUGGUGUU ((.(((((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))(((.((......)).))).........))).)) ( -37.20) >DroYak_CAF1 26912 99 - 1 GAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAUGUUGUUGC---------UGUU ......((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))(((.((......)).)))..---------.... ( -33.80) >DroAna_CAF1 27493 99 - 1 GAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAUGUUGUUGC---------UGUU ......((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))(((.((......)).)))..---------.... ( -33.80) >DroPer_CAF1 29036 98 - 1 GAGACCGUGGGAAACAGAUUUGUCGUGUGU-UCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAUGUUGUUGC---------UGUU ......((((((((((((....)).))).)-)))))).(((((((((.((.((......)).)).)).)))))))(((.((......)).)))..---------.... ( -29.40) >consensus GAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUGCUGAAAUGUUGUUGC_________UGUU ......((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))(((.((......)).)))............... (-33.33 = -33.50 + 0.17)


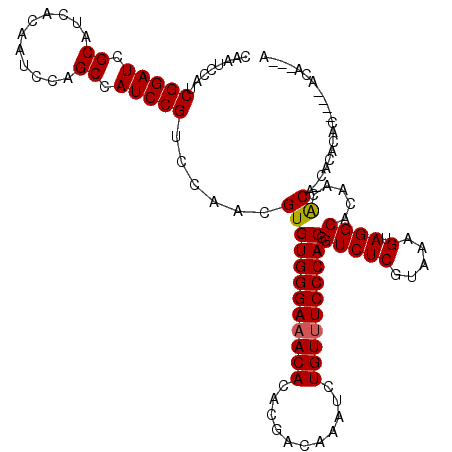
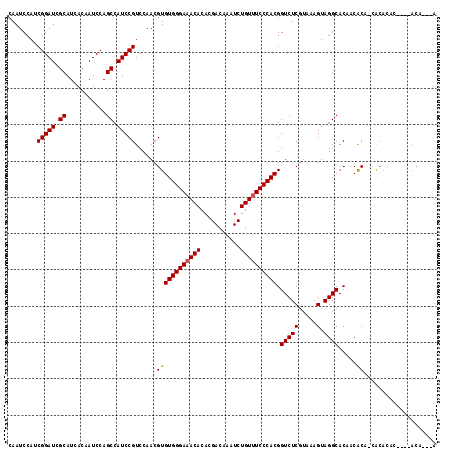
| Location | 21,846,430 – 21,846,539 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 90.39 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -24.83 |
| Energy contribution | -24.75 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21846430 109 + 27905053 CAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACACCGCA-CACACAC----AC----A ....((..(((((.((...........)).)))))........((((((((((...........)))))))))))).........((..((.....)).-.))....----..----. ( -25.60) >DroSec_CAF1 29935 113 + 1 CAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACACCGCA-CACACAC----ACACAGA ....((..(((((.((...........)).)))))........((((((((((...........)))))))))))).........((..((.....)).-.))....----....... ( -25.60) >DroEre_CAF1 23339 115 + 1 CAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACACCGCA-CACACACAC--ACACACA ....((..(((((.((...........)).)))))........((((((((((...........)))))))))))).........((..((.....)).-.))......--....... ( -25.60) >DroMoj_CAF1 28441 108 + 1 CAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACAACACA--ACACAC----ACU---- ........(((((.((...........)).))))).......(((((((((((...........)))))))))))(((((.....).))))........--......----...---- ( -25.00) >DroAna_CAF1 27512 118 + 1 CAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACAACACAGCGCAACUAUACACACACA ....((..(((((.((...........)).)))))........((((((((((...........))))))))))))....(((.(((.(((........)).).))).)))....... ( -29.00) >DroPer_CAF1 29055 100 + 1 CAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGA-ACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACCACACA-CA---------------- ........(((((.((...........)).)))))......((((((((-(.(((.((....))))))))))))(((.((........)).)))))...-..---------------- ( -23.60) >consensus CAAUCCAUCGGAUCGCAUCACAAUCCAGCCAUCCGUCCAACGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACAACACA_CACACAC____ACA___A ........(((((.((...........)).)))))......((((((((((((...........)))))))))).(((((.....).)))).....)).................... (-24.83 = -24.75 + -0.08)

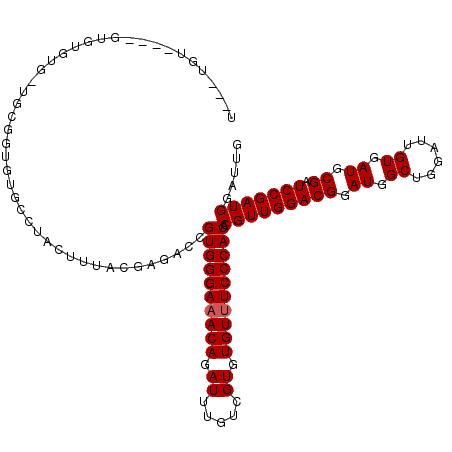
| Location | 21,846,430 – 21,846,539 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 90.39 |
| Mean single sequence MFE | -38.38 |
| Consensus MFE | -32.63 |
| Energy contribution | -32.80 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21846430 109 - 27905053 U----GU----GUGUGUG-UGCGGUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUG .----.(----(((...(-((.((....)))))..)))).....((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))..... ( -38.00) >DroSec_CAF1 29935 113 - 1 UCUGUGU----GUGUGUG-UGCGGUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUG (((.(((----(...(((-.((.....)).)))..)))))))..((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))..... ( -39.10) >DroEre_CAF1 23339 115 - 1 UGUGUGU--GUGUGUGUG-UGCGGUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUG .(.((.(--.((((...(-((.((....)))))..))))).)))((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))..... ( -38.80) >DroMoj_CAF1 28441 108 - 1 ----AGU----GUGUGU--UGUGUUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUG ----...----(.((.(--((((..((....))..))))).)))((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))..... ( -39.50) >DroAna_CAF1 27512 118 - 1 UGUGUGUGUAUAGUUGCGCUGUGUUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUG .(.((.((((.(((.((((......))))..))).))))..)))((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))..... ( -43.70) >DroPer_CAF1 29055 100 - 1 ----------------UG-UGUGUGGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGU-UCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUG ----------------.(-((.((((...))))..)))......((((((((((((....)).))).)-)))))).(((((((((.((.((......)).)).)).)))))))..... ( -31.20) >consensus U___UGU____GUGUGUG_UGCGGUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACGUUGGACGGAUGGCUGGAUUGUGAUGCGAUCCGAUGGAUUG ............................................((((((((((.((.....)).)))))))))).(((((((((.((.((......)).)).)).)))))))..... (-32.63 = -32.80 + 0.17)


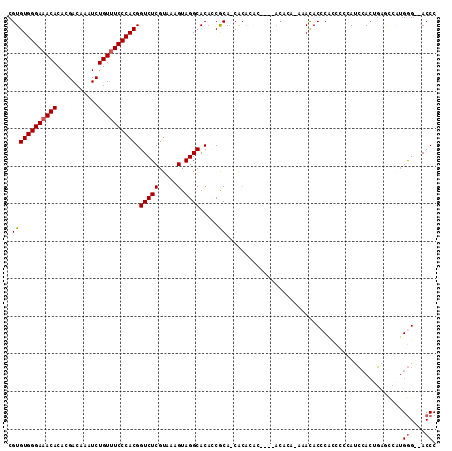
| Location | 21,846,470 – 21,846,573 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.45 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -17.86 |
| Energy contribution | -18.02 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21846470 103 + 27905053 CGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACACCGCA-CACACAC----AC----AAACACCCAUCCCCAUCCACUGAGCCAUGGG--ACCC ...((((((((((...........))))))))))((((((((...((..((.....)).-.))....----..----.............((.....))....)))))--))). ( -26.60) >DroSec_CAF1 29975 101 + 1 CGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACACCGCA-CACACAC----ACACAGAGACACCC------AUCCACUGAGCCAUGGG--ACCC ...((((((((((...........))))))))))((((((((...((..((.....)).-.))....----...(((.((.....------.))..)))....)))))--))). ( -29.20) >DroEre_CAF1 23379 109 + 1 CGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACACCGCA-CACACACAC--ACACACAAACACCCAUCCCCAACCACUGAACCAUGGG--ACCC ...((((((((((...........))))))))))((((((((...((..((.....)).-.))......--...................((.....))....)))))--))). ( -26.80) >DroAna_CAF1 27552 114 + 1 CGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACAACACAGCGCAACUAUACACACACAGAUACACACUGACACCGAGGGACUCUUGGGAGACCC ...((((((((((...........))))))))))(((((((((.(((.(((........)).).))).))).....(((.......)))...(((((.....))))))))))). ( -37.20) >DroPer_CAF1 29095 91 + 1 CGUGUGGGA-ACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACCACACA-CA----------------CACACACUCUCGCAUGCACCCAG---AGAG--ACCC ...((((((-(.(((.((....))))))))))))((((((.....(((.((........-..----------------...........)).)))......---.)))--))). ( -23.40) >consensus CGUGUGGGAAACACACGACAAAUCUGUUUCCCACGGUCUCGUAAAGUAGGCACACCGCA_CACACAC____ACACA_AAACACCCACCCCCAUCCACUGAGCCAUGGG__ACCC .((((((((((((...........)))))))))).(((((.....).)))).....))...............................................((....)). (-17.86 = -18.02 + 0.16)

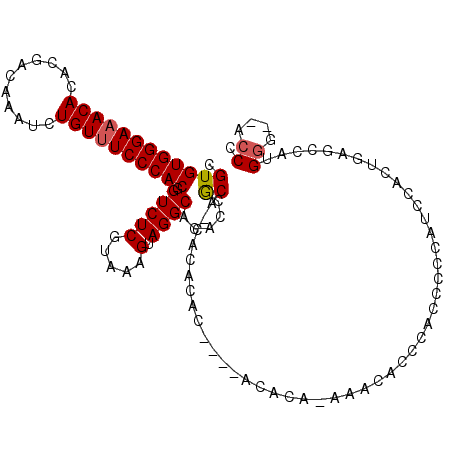
| Location | 21,846,470 – 21,846,573 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.45 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -20.89 |
| Energy contribution | -20.77 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.869947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21846470 103 - 27905053 GGGU--CCCAUGGCUCAGUGGAUGGGGAUGGGUGUUU----GU----GUGUGUG-UGCGGUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACG .(((--(((((......)))).(((((.((((..(((----(.----.(....)-..))).)..)))))))))...))))((((((((((.((.....)).))))))))))... ( -37.20) >DroSec_CAF1 29975 101 - 1 GGGU--CCCAUGGCUCAGUGGAU------GGGUGUCUCUGUGU----GUGUGUG-UGCGGUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACG (..(--((((((((((.((((((------(((..(..(((..(----......)-..))).)..)))).)))))))).))))))))..)........((((((.....)))))) ( -41.60) >DroEre_CAF1 23379 109 - 1 GGGU--CCCAUGGUUCAGUGGUUGGGGAUGGGUGUUUGUGUGU--GUGUGUGUG-UGCGGUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACG .(((--(((((......)))).(((((.((((..((((((..(--......)..-))))).)..)))))))))...))))((((((((((.((.....)).))))))))))... ( -37.60) >DroAna_CAF1 27552 114 - 1 GGGUCUCCCAAGAGUCCCUCGGUGUCAGUGUGUAUCUGUGUGUGUAUAGUUGCGCUGUGUUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACG (((.(((....))).)))..(((.((...(((((.(((((....))))).))))).(((..((....))..))))).)))((((((((((.((.....)).))))))))))... ( -43.30) >DroPer_CAF1 29095 91 - 1 GGGU--CUCU---CUGGGUGCAUGCGAGAGUGUGUG----------------UG-UGUGUGGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGU-UCCCACACG .(((--(((.---....(..(((((....)))))..----------------)(-((.((((...))))..)))))))))((((((((((((....)).))).)-))))))... ( -35.20) >consensus GGGU__CCCAUGGCUCAGUGGAUGGGAAUGGGUGUCU_UGUGU____GUGUGUG_UGCGGUGUGCCUACUUUACGAGACCGUGGGAAACAGAUUUGUCGUGUGUUUCCCACACG ((((....(((.((..........................................)).))).)))).............((((((((((.((.....)).))))))))))... (-20.89 = -20.77 + -0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:18 2006