
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,834,160 – 21,834,273 |
| Length | 113 |
| Max. P | 0.887273 |

| Location | 21,834,160 – 21,834,251 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -14.30 |
| Energy contribution | -14.30 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
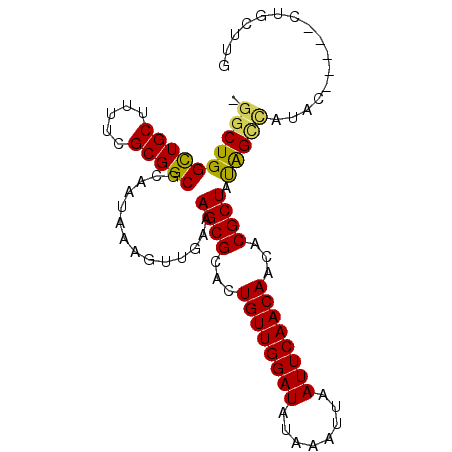
>3R_DroMel_CAF1 21834160 91 + 27905053 -------------AGAGACCCAGAGCAACAACACCAAGCAGUGUGUAUGGCCAUAGCGUGUUGUUGAAUUAAUUUAUAUCCAACAGUGCGCUUUCAACUUUAUU -------------.((((....(.((....((((......)))).....)))...(((..((((((.((........)).))))))..)))))))......... ( -21.00) >DroPse_CAF1 15578 88 + 1 -------------AGAGACCCAGAGAUACAGCAACAACCAC---ACACAACUGUAGCGUGUUGUUGAAUUAAUUUAUAUCCAACAGUGCGCUUUCAACUUUAUU -------------.........(((((((((..........---......)))))(((..((((((.((........)).))))))..)))))))......... ( -17.99) >DroEre_CAF1 10868 88 + 1 ----------------GACCCAGAGCAACAACACCAAGCAGUGUGUAUGGCCAUAGCGUGUUGUUGAAUUAAUUUAUAUCCAACAGUGCGCUUUCAACUUUAUU ----------------......(.((....((((......)))).....)))..((((..((((((.((........)).))))))..))))............ ( -19.80) >DroYak_CAF1 11051 104 + 1 GGAGACCGGAGACGGAGACCCAGAGCAACAACACCAAGCUGUGUGUAUGGCCAUAGCGUGUUGUUGAAUUAAUUUAUAUCCAACAGUGCGCUUUCAACUUUAUU ((((.(((....)))......(((((...........((((((.(.....)))))))(..((((((.((........)).))))))..))))))...))))... ( -29.50) >DroAna_CAF1 14266 71 + 1 ----------------GACCCAGAGCAACACAA-----------------CUAUAGCGUGUUGUUGAAUUAAUUUAUAUCCAACAGUGCGCUUUCAACUUUAUU ----------------.................-----------------....((((..((((((.((........)).))))))..))))............ ( -14.30) >DroPer_CAF1 15288 88 + 1 -------------AGAGACCCAGAGAUACAGCAACAACCAC---ACACAACUGUAGCGUGUUGUUGAAUUAAUUUAUAUCCAACAGUGCGCUUUCAACUUUAUU -------------.........(((((((((..........---......)))))(((..((((((.((........)).))))))..)))))))......... ( -17.99) >consensus _____________AGAGACCCAGAGCAACAACACCAAGCAG___GUAUGGCCAUAGCGUGUUGUUGAAUUAAUUUAUAUCCAACAGUGCGCUUUCAACUUUAUU ......................................................((((..((((((.((........)).))))))..))))............ (-14.30 = -14.30 + -0.00)



| Location | 21,834,181 – 21,834,273 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 86.98 |
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -23.99 |
| Energy contribution | -23.30 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.29 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21834181 92 - 27905053 -GGCUGGCUGCUUUUCGCGGCGCAAUAAAGUUGAAAGCGCACUGUUGGAUAUAAAUUAAUUCAACAACACGCUAUGGCCAUACACA--CUGCUUG -((((.(((((.....)))))..............((((...((((((((........))))))))...))))..)))).......--....... ( -25.90) >DroPse_CAF1 15599 90 - 1 GGGCUGGUUGCUUUUCGCGGCGCAAUAAAGUUGAAAGCGCACUGUUGGAUAUAAAUUAAUUCAACAACACGCUACAGUUGUGU-----GUGGUUG (.(((.(((((.....))))).((((...))))..))).)..((((((((........))))))))((((((.......))))-----))..... ( -23.10) >DroSim_CAF1 11112 94 - 1 -GGCUGGCUGCUUUUCGCGGCGCAAUAAAGUUGAAAGCGCACUGUUGGAUAUAAAUUAAUUCAACAACACGCUAUGGCCAUACACACUCUGCUUG -((((.(((((.....)))))..............((((...((((((((........))))))))...))))..))))................ ( -25.90) >DroYak_CAF1 11085 92 - 1 -GGCUGGCUGCUUUUCGCGGCGCAAUAAAGUUGAAAGCGCACUGUUGGAUAUAAAUUAAUUCAACAACACGCUAUGGCCAUACACA--CAGCUUG -((((((((((.....)))))..............((((...((((((((........))))))))...)))).............--))))).. ( -26.10) >DroAna_CAF1 14283 76 - 1 -GGCUGGCUGCUUUUCGCGGCGCAAUAAAGUUGAAAGCGCACUGUUGGAUAUAAAUUAAUUCAACAACACGCUAUAG------------------ -.((..(((((.....)))))))............((((...((((((((........))))))))...))))....------------------ ( -20.20) >DroPer_CAF1 15309 90 - 1 GGGCUGGUUGCUUUUCGCGGCGCAAUAAAGUUGAAAGCGCACUGUUGGAUAUAAAUUAAUUCAACAACACGCUACAGUUGUGU-----GUGGUUG (.(((.(((((.....))))).((((...))))..))).)..((((((((........))))))))((((((.......))))-----))..... ( -23.10) >consensus _GGCUGGCUGCUUUUCGCGGCGCAAUAAAGUUGAAAGCGCACUGUUGGAUAUAAAUUAAUUCAACAACACGCUAUAGCCAUAC_____CUGCUUG .((((((((((.....)))))..............((((...((((((((........))))))))...)))).)))))................ (-23.99 = -23.30 + -0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:09 2006