| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,770,977 – 2,771,073 |
| Length | 96 |
| Max. P | 0.896079 |

| Location | 2,770,977 – 2,771,073 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.99 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -12.52 |
| Energy contribution | -12.55 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
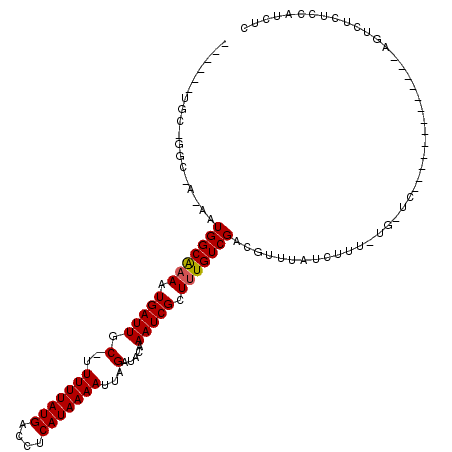
>3R_DroMel_CAF1 2770977 96 + 27905053 ------UGC-GGC-A-AAUGGCGAAAUGAUUGC-UUUUUAUGACCUCAUAAAAUUAGAUACAAAUCGCUUUGUCGACGUUUAUCUUU-UGCUC-------------GGUAUCUCCAUCUC ------..(-(((-(-(..(((((..(((((..-.(((((((....)))))))...))).))..)))))))))))............-.....-------------((.....))..... ( -20.80) >DroPse_CAF1 76856 105 + 1 ------GGC-GGC-AUUAUGGCAAAAUGAUUGCCUUUUUAUGACCUCAUAAAAUUAGAUACAAAUCGCUUUGUCGACGUUUAUCUUUUUGGUC-------GUCUGCCAUUGCUCCAUCUC ------((.-(((-(..((((((....(((.((..(((((((....)))))))...(((....)))))...)))((((.(((......))).)-------)))))))))))))))..... ( -26.60) >DroGri_CAF1 89588 96 + 1 UGAGUCUGGGGCC-A-AAUGGCAAAAUGAUUGC-UUUUUAUGACCUCAUAAAAUUAGAUACAAAUCGCUCUGUCGACAUUUAUCUU---------------------UUCUCUCUCUAUC .(((...((((((-.-...)))....((((.((-.(((((((....)))))))...(((....)))))...))))...........---------------------..))).))).... ( -17.80) >DroWil_CAF1 62382 105 + 1 ------UGC-GGCCA-AAUGGCAAAAUGAUUGC-UUUUUAUGACCUCAUAAAAUUAGAUACAAAUCGUUUUGUCGACAUUUAUCUAU-CGCUAUGUUGUGG-----AGUGUCUUUAGCUU ------.((-.....-...((((((((((((.(-((((((((....)))))))..)).....))))))))))))(((((...(((((-.((...)).))))-----))))))....)).. ( -26.50) >DroMoj_CAF1 96007 97 + 1 UGAGACUGGGGCC-A-AAUGGCAAAAUGAUUGC-UUUUUAUGACCUCAUAAAAUUAGAUACAAAUCGCUUUGUCGACGUUUAUCUC--------------------AGUCUCUCCCUCUC .((((((((((..-(-(.(((((((.(((((.(-((((((((....)))))))..)).....))))).)))))))...))..))))--------------------))))))........ ( -27.90) >DroPer_CAF1 76389 105 + 1 ------GGC-GGC-AUUAUGGCAAAAUGAUUGCCUUUUUAUGACCUCAUAAAAUUAGAUACAAAUCGCUUUGUCGACGUUUAUCUUUUUGGUC-------GUCUGCCAUUGCUCCAUCUC ------((.-(((-(..((((((....(((.((..(((((((....)))))))...(((....)))))...)))((((.(((......))).)-------)))))))))))))))..... ( -26.60) >consensus ______UGC_GGC_A_AAUGGCAAAAUGAUUGC_UUUUUAUGACCUCAUAAAAUUAGAUACAAAUCGCUUUGUCGACGUUUAUCUUU_UG_UC_____________AGUCUCUCCAUCUC ..................(((((((.(((((.(..(((((((....)))))))...).....))))).)))))))............................................. (-12.52 = -12.55 + 0.03)

| Location | 2,770,977 – 2,771,073 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.99 |
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -13.73 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
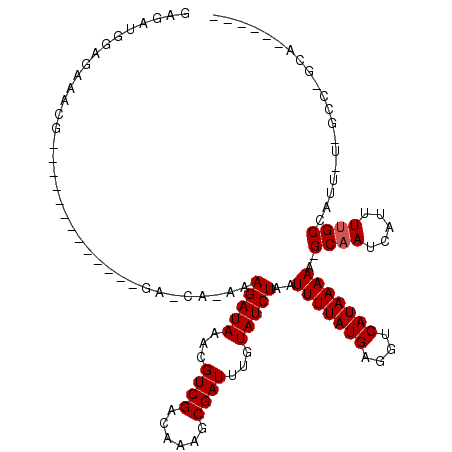
>3R_DroMel_CAF1 2770977 96 - 27905053 GAGAUGGAGAUACC-------------GAGCA-AAAGAUAAACGUCGACAAAGCGAUUUGUAUCUAAUUUUAUGAGGUCAUAAAAA-GCAAUCAUUUCGCCAUU-U-GCC-GCA------ .((((((.((....-------------..((.-..(((((...((((......))))...)))))..(((((((....))))))).-)).......)).)))))-)-...-...------ ( -20.42) >DroPse_CAF1 76856 105 - 1 GAGAUGGAGCAAUGGCAGAC-------GACCAAAAAGAUAAACGUCGACAAAGCGAUUUGUAUCUAAUUUUAUGAGGUCAUAAAAAGGCAAUCAUUUUGCCAUAAU-GCC-GCC------ .............((((...-------((((....(((((...((((......))))...)))))..((....)))))).......(((((.....)))))....)-)))-...------ ( -25.10) >DroGri_CAF1 89588 96 - 1 GAUAGAGAGAGAA---------------------AAGAUAAAUGUCGACAGAGCGAUUUGUAUCUAAUUUUAUGAGGUCAUAAAAA-GCAAUCAUUUUGCCAUU-U-GGCCCCAGACUCA ....(((.((((.---------------------.(((((...((((......))))...)))))..)))).((.(((((......-((((.....))))....-)-)))).))..))). ( -21.90) >DroWil_CAF1 62382 105 - 1 AAGCUAAAGACACU-----CCACAACAUAGCG-AUAGAUAAAUGUCGACAAAACGAUUUGUAUCUAAUUUUAUGAGGUCAUAAAAA-GCAAUCAUUUUGCCAUU-UGGCC-GCA------ ..((..........-----......(((((.(-.((((((...((((......))))...)))))).).))))).(((((......-((((.....))))....-)))))-)).------ ( -23.70) >DroMoj_CAF1 96007 97 - 1 GAGAGGGAGAGACU--------------------GAGAUAAACGUCGACAAAGCGAUUUGUAUCUAAUUUUAUGAGGUCAUAAAAA-GCAAUCAUUUUGCCAUU-U-GGCCCCAGUCUCA ........((((((--------------------((((((...((((......))))...)))))..........(((((......-((((.....))))....-)-)))).))))))). ( -28.10) >DroPer_CAF1 76389 105 - 1 GAGAUGGAGCAAUGGCAGAC-------GACCAAAAAGAUAAACGUCGACAAAGCGAUUUGUAUCUAAUUUUAUGAGGUCAUAAAAAGGCAAUCAUUUUGCCAUAAU-GCC-GCC------ .............((((...-------((((....(((((...((((......))))...)))))..((....)))))).......(((((.....)))))....)-)))-...------ ( -25.10) >consensus GAGAUGGAGAAACG_____________GA_CA_AAAGAUAAACGUCGACAAAGCGAUUUGUAUCUAAUUUUAUGAGGUCAUAAAAA_GCAAUCAUUUUGCCAUU_U_GCC_GCA______ ...................................(((((...((((......))))...)))))..(((((((....)))))))..((((.....)))).................... (-13.73 = -13.90 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:47 2006