
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,811,685 – 21,811,816 |
| Length | 131 |
| Max. P | 0.889559 |

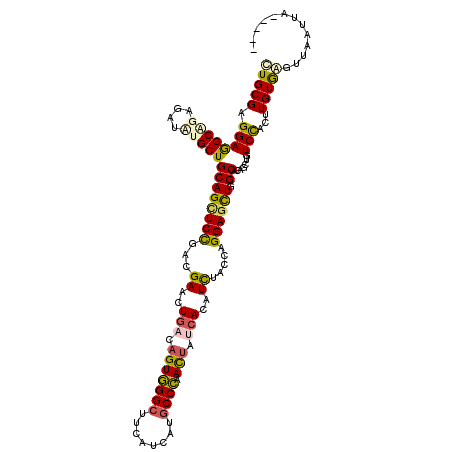
| Location | 21,811,685 – 21,811,798 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -38.55 |
| Consensus MFE | -26.13 |
| Energy contribution | -26.00 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.739173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21811685 113 + 27905053 AUGCGAGGAGGCUGAGAUUUGCUGCAGUUCUGAUGAAUUGACAGUCGGAUUUAUUAUGCCGAAUUACCACAUUUACCAGGAGCUGGCUCAGUUGUCCACAUGUGAGUUUGGUA----- .........(((..((((((((((((((((....)))))).))).))))))).....)))....(((((.((((((..(((((((...))))..)))....)))))).)))))----- ( -32.70) >DroVir_CAF1 112191 118 + 1 UUGCGAGGAGGCAGAGAUCUGCUGCAGCUCCGAUGAGUUGACCGUGGGCUUCAUCAUGCCCAACUAUCACAUCUACCAGGAACUGGCGCAGCUCUCUUCUUGUGAGUUAUUUGGGAUU (..(((((((..((((..((((..(((.(((...((..(((...(((((........)))))....)))..)).....))).)))..)))))))))))))))..)............. ( -37.80) >DroPse_CAF1 87448 113 + 1 CUGCGAGGAGGCAGAGAUAUGCUGCAGCUCCGACGAACUGACAGUGGGCUUCAUUAUGCCCAACUAUCACAUCUACCAGGAGCUGGCACAGUUGUCCACUUGUGGGUUGAUUA----- (..((((..(((((...(.(((..(((((((...((..(((.(((((((........)))).))).)))..)).....)))))))))).).)))))..))))..)........----- ( -41.30) >DroMoj_CAF1 117478 118 + 1 CUGCGAGGAAGCGGAGAUCUGCUGCAGCUCCGAUGAGUUAACAGUGGGCUUCAUCAUGCCCAACUACCACAUUUACCAGGAGCUAGCCCAACUCUCUUCAUGUGAGUUAAAUGCGAUU .((((.((.(((((....)))))((((((((..(((((....(((((((........)))).))).....)))))...)))))).))))(((((.(.....).)))))...))))... ( -38.30) >DroAna_CAF1 81540 108 + 1 UUGCGAGGAAGCUGAGAUUUGCUGCAGCUCUGACGAGCUGACAGUGGGCUUCAUCAUGCCCAACUAUCACAUUUACCAGGAGUUGGCACAGCUGUCCUCCUGUAAGUU---------- ((((((((((((((.(((..((((((((((....)))))).))))((((........))))....))).......((((...))))..))))).)))))..))))...---------- ( -39.90) >DroPer_CAF1 87843 113 + 1 CUGCGAGGAGGCAGAGAUAUGCUGCAGCUCCGACGAACUGACAGUGGGCUUCAUUAUGCCCAACUAUCACAUCUACCAGGAGCUGGCACAGUUGUCCACUUGUGGGUUGAUUA----- (..((((..(((((...(.(((..(((((((...((..(((.(((((((........)))).))).)))..)).....)))))))))).).)))))..))))..)........----- ( -41.30) >consensus CUGCGAGGAGGCAGAGAUAUGCUGCAGCUCCGACGAACUGACAGUGGGCUUCAUCAUGCCCAACUAUCACAUCUACCAGGAGCUGGCACAGCUGUCCACUUGUGAGUUAAUUA_____ (((((.((((((((....)))))((((((((...((..(((.(((((((........)))).))).)))..)).....)))))).)).......)))...)))))............. (-26.13 = -26.00 + -0.13)


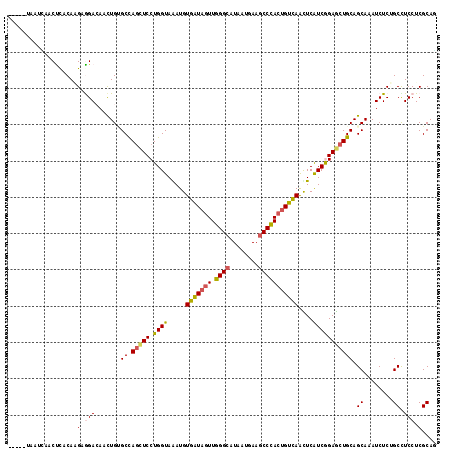
| Location | 21,811,685 – 21,811,798 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -23.07 |
| Energy contribution | -23.02 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21811685 113 - 27905053 -----UACCAAACUCACAUGUGGACAACUGAGCCAGCUCCUGGUAAAUGUGGUAAUUCGGCAUAAUAAAUCCGACUGUCAAUUCAUCAGAACUGCAGCAAAUCUCAGCCUCCUCGCAU -----............(((((((...(((((((((...))))....((..((...((((.((.....))))))(((.........))).))..))......)))))..)))..)))) ( -21.10) >DroVir_CAF1 112191 118 - 1 AAUCCCAAAUAACUCACAAGAAGAGAGCUGCGCCAGUUCCUGGUAGAUGUGAUAGUUGGGCAUGAUGAAGCCCACGGUCAACUCAUCGGAGCUGCAGCAGAUCUCUGCCUCCUCGCAA .....................(((((.((((..(((((((.(((.((..((((...(((((........)))))..))))..))))))))))))..)))).)))))((......)).. ( -39.80) >DroPse_CAF1 87448 113 - 1 -----UAAUCAACCCACAAGUGGACAACUGUGCCAGCUCCUGGUAGAUGUGAUAGUUGGGCAUAAUGAAGCCCACUGUCAGUUCGUCGGAGCUGCAGCAUAUCUCUGCCUCCUCGCAG -----........(((....))).....((((((((((((.((..(((.(((((((.((((........))))))))))))))..)))))))))..)))))...((((......)))) ( -41.70) >DroMoj_CAF1 117478 118 - 1 AAUCGCAUUUAACUCACAUGAAGAGAGUUGGGCUAGCUCCUGGUAAAUGUGGUAGUUGGGCAUGAUGAAGCCCACUGUUAACUCAUCGGAGCUGCAGCAGAUCUCCGCUUCCUCGCAG ....((..(((((((.(.....).)))))))((.((((((.(((.....(..((((.((((........))))))))..)....))))))))))).))........((......)).. ( -34.90) >DroAna_CAF1 81540 108 - 1 ----------AACUUACAGGAGGACAGCUGUGCCAACUCCUGGUAAAUGUGAUAGUUGGGCAUGAUGAAGCCCACUGUCAGCUCGUCAGAGCUGCAGCAAAUCUCAGCUUCCUCGCAA ----------.........(((((.((((((((((.....)))))..(((((((((.((((........))))))))))(((((....)))))...))).....)))))))))).... ( -42.30) >DroPer_CAF1 87843 113 - 1 -----UAAUCAACCCACAAGUGGACAACUGUGCCAGCUCCUGGUAGAUGUGAUAGUUGGGCAUAAUGAAGCCCACUGUCAGUUCGUCGGAGCUGCAGCAUAUCUCUGCCUCCUCGCAG -----........(((....))).....((((((((((((.((..(((.(((((((.((((........))))))))))))))..)))))))))..)))))...((((......)))) ( -41.70) >consensus _____UAAUCAACUCACAAGAGGACAACUGUGCCAGCUCCUGGUAAAUGUGAUAGUUGGGCAUAAUGAAGCCCACUGUCAACUCAUCGGAGCUGCAGCAAAUCUCUGCCUCCUCGCAG ...................(.(((.......((.(((((.((((.....(((((((.((((........)))))))))))....))))))))))).((........)).))).).... (-23.07 = -23.02 + -0.05)


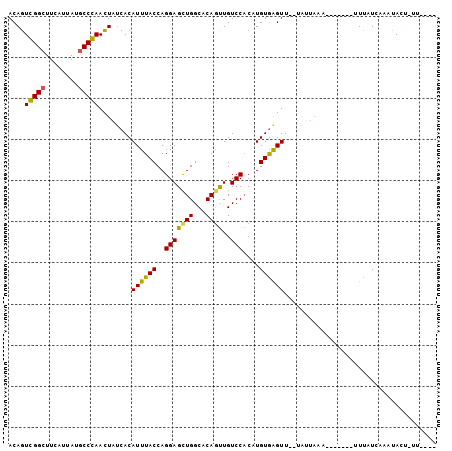
| Location | 21,811,725 – 21,811,816 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.11 |
| Mean single sequence MFE | -21.22 |
| Consensus MFE | -14.71 |
| Energy contribution | -13.72 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21811725 91 + 27905053 ACAGUCGGAUUUAUUAUGCCGAAUUACCACAUUUACCAGGAGCUGGCUCAGUUGUCCACAUGUGAGUU--UGGUAAA-------UUCAUCAUACAUAUUU---- ....((((((.....)).)))).((((((.((((((..(((((((...))))..)))....)))))).--)))))).-------................---- ( -18.60) >DroPse_CAF1 87488 95 + 1 ACAGUGGGCUUCAUUAUGCCCAACUAUCACAUCUACCAGGAGCUGGCACAGUUGUCCACUUGUGGGUU--GAUUAAAGCAUUUGUUUAUCAAGUAAU------- ..(((((((........)))).)))..........(((.(((.(((.((....)))))))).))).((--(((..((((....))))))))).....------- ( -24.70) >DroEre_CAF1 87598 93 + 1 ACAGUCGGCUUUAUUAUGCCGAAUUAUCACAUUUACCAGGAGCUGGCUCAGUUGUCCACAUGUGAGUUUUUAUUAAA-------UUCAUAAUCUACGCUU---- ....(((((........))))).................((((((((......).)))..(((((((((.....)))-------))))))......))))---- ( -20.20) >DroYak_CAF1 79553 91 + 1 ACAGUCGGCUUUAUUAUGCCGAAUUAUCACAUUUACCAGGAGCUGGCUCAAUUGUCCACAUGUGAGUU--UGUUAAA-------UUUAUCAUCUACUCUU---- ....(((((........)))))..................(((..(((((..((....))..))))).--.)))...-------................---- ( -17.20) >DroAna_CAF1 81580 90 + 1 ACAGUGGGCUUCAUCAUGCCCAACUAUCACAUUUACCAGGAGUUGGCACAGCUGUCCUCCUGUAAGUU-------AA-------UUUAUAAAACACUAUUUCAA ..(((((((........)))).))).....((((((.(((((..((((....))))))))))))))).-------..-------.................... ( -21.90) >DroPer_CAF1 87883 95 + 1 ACAGUGGGCUUCAUUAUGCCCAACUAUCACAUCUACCAGGAGCUGGCACAGUUGUCCACUUGUGGGUU--GAUUAAAGCAUUUGUUUAUCAAGUAAU------- ..(((((((........)))).)))..........(((.(((.(((.((....)))))))).))).((--(((..((((....))))))))).....------- ( -24.70) >consensus ACAGUCGGCUUCAUUAUGCCCAACUAUCACAUUUACCAGGAGCUGGCACAGUUGUCCACAUGUGAGUU__UAUUAAA_______UUUAUCAAAUACU_UU____ ....(((((........)))))........((((((..(((((((...))))..)))....))))))..................................... (-14.71 = -13.72 + -1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:56 2006