
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,768,412 – 21,768,527 |
| Length | 115 |
| Max. P | 0.514928 |

| Location | 21,768,412 – 21,768,527 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -41.48 |
| Consensus MFE | -25.17 |
| Energy contribution | -27.38 |
| Covariance contribution | 2.22 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21768412 115 + 27905053 CUUUUCCAUCUGACACAUUCAAUUUGGUCAGGCACAGCUGCUCAGCGGGUUAGUUGCCAUUUUCCUGGGCUUUUUCAGGCGAGGGGGAGGAAGAUGAGCGGCACCACAUGCCGCA----- .((((((.((((((.((.......))))))))..((((((((.....)).))))))...((..(((.(.((.....)).).)))..))))))))...((((((.....)))))).----- ( -40.60) >DroSec_CAF1 35234 115 + 1 CUUUUCCAUCUGACACAUUCAAUUUGGUCGAGCGCAGCUGCUCGGCGGGAUAGUUGCCAUAUUCCUGGGUUUUUCCAGGGGAGGGGGAGGAAGAUGAGCGGCACCACAUGCCGCA----- ......(((((..(.......((((.((((((((....)))))))).))))..((.((...((((((((....))))))))..)).)))..))))).((((((.....)))))).----- ( -47.00) >DroSim_CAF1 36420 115 + 1 CUUUUCCAUCUGACACAUUCAAUUUGGUCGAGCGCAGCUGCUCGACGGGAUAGUUGCCAUUUUCCUGGGUUUUUCCAGGGGAGGGGGAGGAAGAUGAGCGGCACCACAUGCCGCA----- ......(((((..(.......((((.((((((((....)))))))).))))..((.((.((..((((((....))))))..)))).)))..))))).((((((.....)))))).----- ( -49.90) >DroEre_CAF1 43816 115 + 1 CUUUUCCAUCUGACACAUUCAAUUUGGUGGAGCCCAGCUGCUCGGCGGGAUAGUUGCCAUUUUCCUGCGCUUUUCCAGGGGAGGGGGCGGAAGAUGAGCGGCACCACAUGUCGCU----- ...........((((((.......))((((..(((.(((....))))))...(((((((((((((.((.((((((....)))))).)))))))))).))))).)))).))))...----- ( -45.00) >DroYak_CAF1 35826 115 + 1 CUUUUCCAUCUGACACAUUCAAUUUGGUCGAGCGCAGCUGCUCAGCGGGAUAGUUGCCAUUUUCCUGGGCUUUUCCAGCGGAGGGGGAGGAAGAUGAGCGGCACCACAUGUCGAU----- .......(((.((((......((((.((.(((((....))))).)).)))).((((((((((((((...(((((((...))))))).))))))))).)))))......)))))))----- ( -37.30) >DroPer_CAF1 41307 99 + 1 CUUUCUUUUCUGACACAUUCAAUUUGGUCACCCACAGCAGCUUUGUGGGGCAAGCGGCA-------------------GGAAGCAGGUGGA--UUGAGAAGCACCACAUGUUGCCACUUG ..........((((.((.......))))))(((((((.....))))))).((((.((((-------------------....(((.((((.--..........)))).))))))).)))) ( -29.10) >consensus CUUUUCCAUCUGACACAUUCAAUUUGGUCGAGCGCAGCUGCUCGGCGGGAUAGUUGCCAUUUUCCUGGGCUUUUCCAGGGGAGGGGGAGGAAGAUGAGCGGCACCACAUGCCGCA_____ ......(((((..(.......((((.((.((((......)))).)).))))..((.((.((((((((((....)))))))))))).)))..))))).((((((.....))))))...... (-25.17 = -27.38 + 2.22)

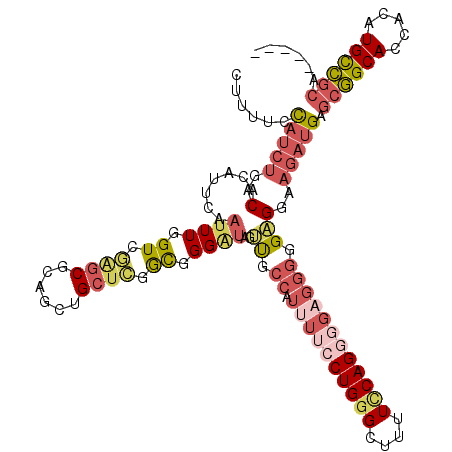
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:43 2006