
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,737,398 – 21,737,511 |
| Length | 113 |
| Max. P | 0.866582 |

| Location | 21,737,398 – 21,737,511 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 66.56 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -12.76 |
| Energy contribution | -12.66 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
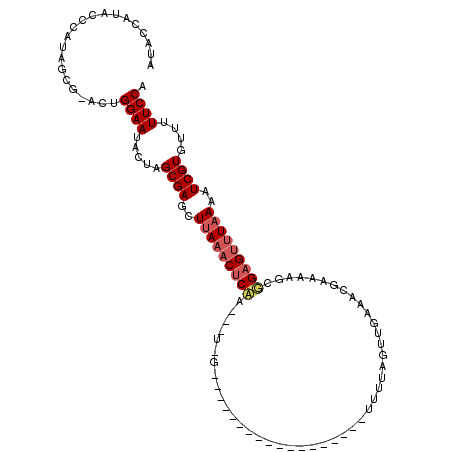
>3R_DroMel_CAF1 21737398 113 + 27905053 UGGAAAAACACGAUUUUAAACUCCGCUUAUCGUUUCAACUAUAUAUAGAAUAUAAUUUAUAGCGAUUUUUGAGUUUAAGCUCGCUAGUAUUCCGGU-CACUAUGGGUAUGGUAU .((((.....(((.(((((((((.....((((((..((.(((((.....))))).))...))))))....))))))))).)))......))))...-.(((((....))))).. ( -22.40) >DroSim_CAF1 4291 89 + 1 UGGAAAAACACGAUUUUAAACUCCGCUUUUCGUUUCAACUAAAA-------------------------GGAGUUUAAGCUCGCUAGUAUUCCAAACCCCUUUGGGUAUGGUAU (((((.....(((.((((((((((...(((.((....)).))).-------------------------)))))))))).)))......))))).((((....))))....... ( -22.70) >DroEre_CAF1 12496 95 + 1 UGGAAACACACGAUUUAAGGCUCGGCUUUCAGUUUCAAUUAAUA-----------------CC-AAAGUCGGGUUUAAUUUCGCUAGUAUUCCAGU-CGCUAUGUCUAUGAUAA (((((((...(((..(.((((((((((((..(((......))).-----------------..-)))))))))))).)..)))...)).)))))..-................. ( -20.30) >consensus UGGAAAAACACGAUUUUAAACUCCGCUUUUCGUUUCAACUAAAA__________________C_A___UCGAGUUUAAGCUCGCUAGUAUUCCAGU_CACUAUGGGUAUGGUAU (((((.....(((..((((((((((...........................................))))))))))..)))......))))).................... (-12.76 = -12.66 + -0.10)



| Location | 21,737,398 – 21,737,511 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 66.56 |
| Mean single sequence MFE | -21.93 |
| Consensus MFE | -11.30 |
| Energy contribution | -12.75 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21737398 113 - 27905053 AUACCAUACCCAUAGUG-ACCGGAAUACUAGCGAGCUUAAACUCAAAAAUCGCUAUAAAUUAUAUUCUAUAUAUAGUUGAAACGAUAAGCGGAGUUUAAAAUCGUGUUUUUCCA .................-...((((.....((((..((((((((....((((.....((((((((.....))))))))....)))).....))))))))..))))....)))). ( -26.50) >DroSim_CAF1 4291 89 - 1 AUACCAUACCCAAAGGGGUUUGGAAUACUAGCGAGCUUAAACUCC-------------------------UUUUAGUUGAAACGAAAAGCGGAGUUUAAAAUCGUGUUUUUCCA ...(((.((((....)))).))).......((((..(((((((((-------------------------((((.((....))..)))).)))))))))..))))......... ( -26.30) >DroEre_CAF1 12496 95 - 1 UUAUCAUAGACAUAGCG-ACUGGAAUACUAGCGAAAUUAAACCCGACUUU-GG-----------------UAUUAAUUGAAACUGAAAGCCGAGCCUUAAAUCGUGUGUUUCCA .......((((((((((-.((((....)))))..((((((..(((....)-))-----------------..))))))..........))(((........))))))))))... ( -13.00) >consensus AUACCAUACCCAUAGCG_ACUGGAAUACUAGCGAGCUUAAACUCAA___U_G__________________UUUUAGUUGAAACGAAAAGCGGAGUUUAAAAUCGUGUUUUUCCA .....................((((.....((((..(((((((((.............................................)))))))))..))))....)))). (-11.30 = -12.75 + 1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:30 2006