
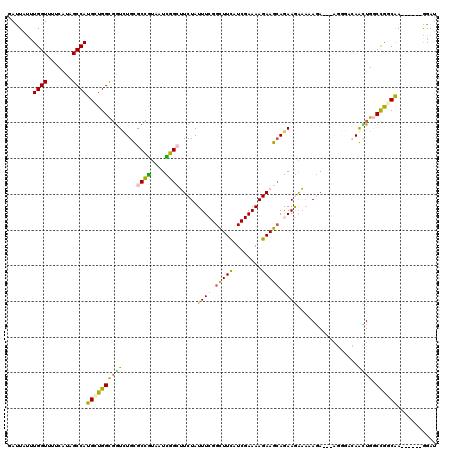
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,733,699 – 21,733,804 |
| Length | 105 |
| Max. P | 0.943992 |

| Location | 21,733,699 – 21,733,804 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.47 |
| Mean single sequence MFE | -36.53 |
| Consensus MFE | -21.78 |
| Energy contribution | -20.82 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
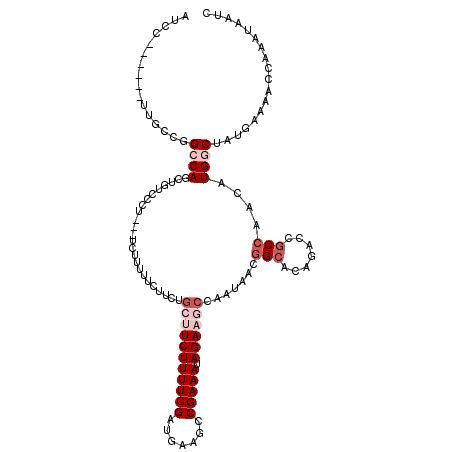
>3R_DroMel_CAF1 21733699 105 + 27905053 GAUUACUUGGUCUUCAUCGCCAUGUUGGCGGUCUGCGCCGUUAUCGGCUUCUAUUUCGGCUUUAUCGAAAAGAAGCAAAAGAAACAGA---------AGCUGGCCGGCAA------GGAC .........(((((.((((((.....))))))....((((...(((((((((.((((.(((((........)))))....)))).)))---------)))))).)))).)------)))) ( -38.30) >DroVir_CAF1 15770 117 + 1 GAUUAUUUGGUUUUCAUAGCCAUGCUGGUGGUCUGUGCCGUAAUUGGCUUCUAUUUCGGCUUCAUCGAAAAGAAGAAGAAAAAGCAAA---AGGGCCAAGUGCCCGCCGAUGAAAAAGAU ........((....(((((((((....)))).)))))))...((((((..((.((((..((((........))))..)))).))....---.((((.....))))))))))......... ( -31.00) >DroPse_CAF1 314 114 + 1 GACUAUUUGGUGUUCAUAGCCAUGCUGGCGGUCUGCGCCGUCAUCGGGGUCUAUUUCGGCUUCAUCGAAAAGAAGCAGAAGGCAAAGAAGAAGGGACAGCUGGCCGGCAA------GGAU .......((((.......))))(((((((.(.((((.((.((.((...((((...((.(((((........))))).))))))...)).)).))).))).).))))))).------.... ( -37.30) >DroGri_CAF1 13077 117 + 1 GAUUAUUUGGUUUUCAUCGCCAUGUUGGCGGUCUGUGCCGUAAUUGGCUUCUAUUUCGGCUUCAUCGAAAAGAAGCAGAAAAAGAAAA---GGGGCCAAUUGUCUACCGAUGAACAAGAU .....((((...(((((((((.....)))(((((.(((((....))))((((.((((.(((((........))))).)))).)))).)---.)))))...........)))))))))).. ( -36.80) >DroWil_CAF1 8138 117 + 1 GAUUAUAUGGUUUUCGUUGCCAUGUUGGCCAUUUGUGCUAUUAUUGGCUUCUAUUUCGGUUUUAUCGAAAAGAAGCAAAAGAAAAAGA---AAUCAAAAUUGGCCAGCAAUGGAACGGAU ............((((((.((((((((((((((((..((.((.((.((((((.(((((((...)))))))))))))...)).)).)).---...))))..)))))))).)))))))))). ( -38.50) >DroPer_CAF1 320 114 + 1 GACUAUUUGGUGUUCAUAGCCAUGCUGGCGGUCUGCGCCGUCAUCGGGGUCUAUUUCGGCUUCAUCGAAAAGAAGCAGAAGGCAAAGAAGAAGGGACAGCUGGCCGGCAA------GGAU .......((((.......))))(((((((.(.((((.((.((.((...((((...((.(((((........))))).))))))...)).)).))).))).).))))))).------.... ( -37.30) >consensus GAUUAUUUGGUUUUCAUAGCCAUGCUGGCGGUCUGCGCCGUAAUCGGCUUCUAUUUCGGCUUCAUCGAAAAGAAGCAGAAGAAAAAGA___AGGGACAACUGGCCGGCAA______GGAU .......((((.......))))((((((((((....((((....)))).....((((.(((((........))))).)))).................)))).))))))........... (-21.78 = -20.82 + -0.96)


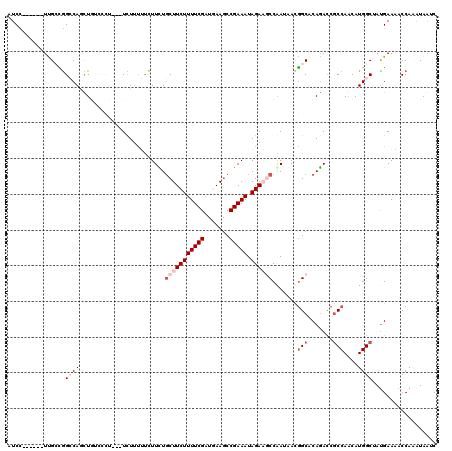
| Location | 21,733,699 – 21,733,804 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.47 |
| Mean single sequence MFE | -30.97 |
| Consensus MFE | -13.33 |
| Energy contribution | -14.83 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21733699 105 - 27905053 GUCC------UUGCCGGCCAGCU---------UCUGUUUCUUUUGCUUCUUUUCGAUAAAGCCGAAAUAGAAGCCGAUAACGGCGCAGACCGCCAACAUGGCGAUGAAGACCAAGUAAUC (((.------.(((((..(.(((---------((((((((....((((..........)))).))))))))))).)....)))))..)))((((.....))))................. ( -33.00) >DroVir_CAF1 15770 117 - 1 AUCUUUUUCAUCGGCGGGCACUUGGCCCU---UUUGCUUUUUCUUCUUCUUUUCGAUGAAGCCGAAAUAGAAGCCAAUUACGGCACAGACCACCAGCAUGGCUAUGAAAACCAAAUAAUC ....(((((((.(((((((.....)))).---..((((.....((((...(((((.......))))).))))(((......)))..........))))..)))))))))).......... ( -28.80) >DroPse_CAF1 314 114 - 1 AUCC------UUGCCGGCCAGCUGUCCCUUCUUCUUUGCCUUCUGCUUCUUUUCGAUGAAGCCGAAAUAGACCCCGAUGACGGCGCAGACCGCCAGCAUGGCUAUGAACACCAAAUAGUC ....------.(((.(((...((((.((.((.((((((..(((.(((((........))))).))).))))....)).)).)).))))...))).))).((((((.........)))))) ( -29.90) >DroGri_CAF1 13077 117 - 1 AUCUUGUUCAUCGGUAGACAAUUGGCCCC---UUUUCUUUUUCUGCUUCUUUUCGAUGAAGCCGAAAUAGAAGCCAAUUACGGCACAGACCGCCAACAUGGCGAUGAAAACCAAAUAAUC ......(((((((.((...(((((((...---..((((.((((.(((((........))))).)))).)))))))))))..(((.......)))....)).)))))))............ ( -34.70) >DroWil_CAF1 8138 117 - 1 AUCCGUUCCAUUGCUGGCCAAUUUUGAUU---UCUUUUUCUUUUGCUUCUUUUCGAUAAAACCGAAAUAGAAGCCAAUAAUAGCACAAAUGGCCAACAUGGCAACGAAAACCAUAUAAUC ...((((((((.(.((((((.....((..---......))....(((((((((((.......))))).))))))...............)))))).))))).)))).............. ( -29.50) >DroPer_CAF1 320 114 - 1 AUCC------UUGCCGGCCAGCUGUCCCUUCUUCUUUGCCUUCUGCUUCUUUUCGAUGAAGCCGAAAUAGACCCCGAUGACGGCGCAGACCGCCAGCAUGGCUAUGAACACCAAAUAGUC ....------.(((.(((...((((.((.((.((((((..(((.(((((........))))).))).))))....)).)).)).))))...))).))).((((((.........)))))) ( -29.90) >consensus AUCC______UUGCCGGCCAGCUGUCCCU___UCUUUUUCUUCUGCUUCUUUUCGAUGAAGCCGAAAUAGAAGCCAAUAACGGCACAGACCGCCAACAUGGCUAUGAAAACCAAAUAAUC ................((((........................(((((((((((.......))))).)))))).......(((.......)))....)))).................. (-13.33 = -14.83 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:28 2006