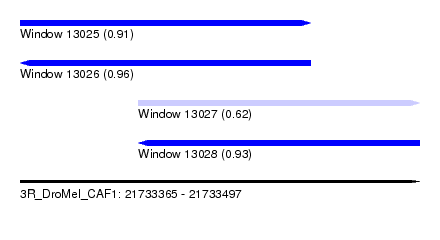
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,733,365 – 21,733,497 |
| Length | 132 |
| Max. P | 0.959513 |

| Location | 21,733,365 – 21,733,461 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 71.89 |
| Mean single sequence MFE | -33.85 |
| Consensus MFE | -15.93 |
| Energy contribution | -18.60 |
| Covariance contribution | 2.67 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
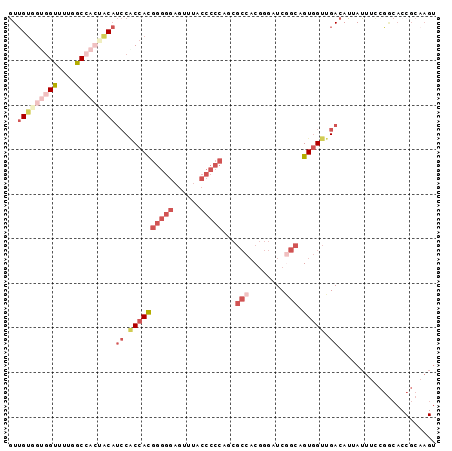
>3R_DroMel_CAF1 21733365 96 + 27905053 ACUUGCGGUGCCGGAAAUAAUGUCAACCACUGCCGAUCCCGUGGCGCUGGGGGUAAACUCCCCCGUGGAGGAUGUAGUGGCCAAAACCACCACAAC ..(((.((((..((..((.(((((..((((.((((......))))...(((((......)))))))))..))))).))..)).....)))).))). ( -35.80) >DroGri_CAF1 12770 81 + 1 ACUUGCGGUCUACAGCAUUAUGUCAACGACUACUGAGGAGG------------UGGCCACAACAAUGGUGGCGGA---CGCUAAUGCUCCAUCUAU ......((.....(((((((((((......((((.....))------------))(((((.......))))).))---)).)))))))))...... ( -21.50) >DroSec_CAF1 2 80 + 1 ----------------AUAAUGUCAACCACUGCCGAUCCCGUGGCGCAGGGGGUAAACUCCCCCGUGGUGGAUGUGGUGGCCAAAACCACCACAAC ----------------.....(((.(((((.((((......))))...(((((......)))))))))).)))(((((((......)))))))... ( -38.30) >DroSim_CAF1 143 96 + 1 ACUUGCGGUGCCGGAAAUAAUGUCAACCACUGCCGAUCCCGUGGCGCAGGGGGUAAACUCCCCCGUGGUGGAUGUAGUGGCCAAAACCACCACAAC ..(((.((((..((..((.(((((.(((((.((((......))))...(((((......)))))))))).))))).))..)).....)))).))). ( -37.10) >DroEre_CAF1 8343 93 + 1 ACUUGCGGUGCCGGAAAUAAUGUCAACCACUGCCGAUCCCGUGGCGUUGGGGGCAAGCUCCCCCGUGGCGUAUGG---UGCCAAAACCGCCACAAC ....((((((...((.......))...)))))).......((((((..(((((......))))).(((((.....---)))))....))))))... ( -35.10) >DroYak_CAF1 143 96 + 1 ACUUGCGGUGCCGGAAAUAAUGUCAACCACAGCGGAACCCGUGGCGUUGGGGGUAAGCUCCCCCGUGGCGGAUGUAGUGGCCAAAACCACCACAAC ..(((.((((..((..((.(((((..((((.((((...))))......(((((......)))))))))..))))).))..)).....)))).))). ( -35.30) >consensus ACUUGCGGUGCCGGAAAUAAUGUCAACCACUGCCGAUCCCGUGGCGCUGGGGGUAAACUCCCCCGUGGUGGAUGUAGUGGCCAAAACCACCACAAC ..........................((((.((((......))))...(((((......)))))))))....(((.((((......)))).))).. (-15.93 = -18.60 + 2.67)

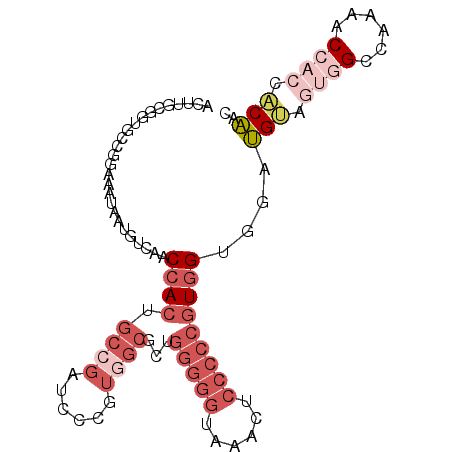
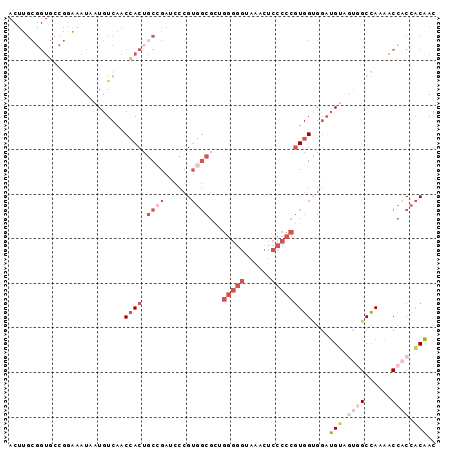
| Location | 21,733,365 – 21,733,461 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 71.89 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -21.92 |
| Energy contribution | -25.00 |
| Covariance contribution | 3.08 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
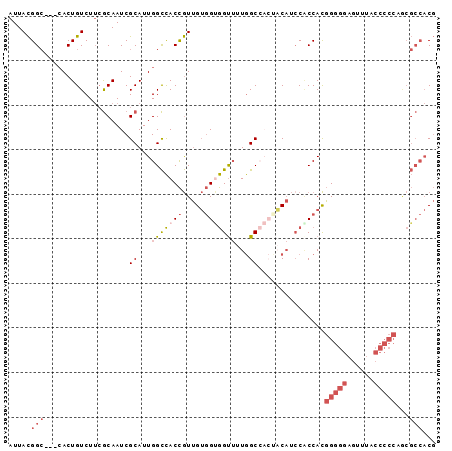
>3R_DroMel_CAF1 21733365 96 - 27905053 GUUGUGGUGGUUUUGGCCACUACAUCCUCCACGGGGGAGUUUACCCCCAGCGCCACGGGAUCGGCAGUGGUUGACAUUAUUUCCGGCACCGCAAGU ..(((((((((....)))))))))((..(((((((((......)))))...(((........))).))))..))....((((.((....)).)))) ( -36.90) >DroGri_CAF1 12770 81 - 1 AUAGAUGGAGCAUUAGCG---UCCGCCACCAUUGUUGUGGCCA------------CCUCCUCAGUAGUCGUUGACAUAAUGCUGUAGACCGCAAGU .....((.(((((((..(---((.((.((.((((..(.((...------------)).)..)))).)).)).))).))))))).))...(....). ( -19.80) >DroSec_CAF1 2 80 - 1 GUUGUGGUGGUUUUGGCCACCACAUCCACCACGGGGGAGUUUACCCCCUGCGCCACGGGAUCGGCAGUGGUUGACAUUAU---------------- ..(((((((((....)))))))))((.((((((((((......)))))...(((........))).))))).))......---------------- ( -38.80) >DroSim_CAF1 143 96 - 1 GUUGUGGUGGUUUUGGCCACUACAUCCACCACGGGGGAGUUUACCCCCUGCGCCACGGGAUCGGCAGUGGUUGACAUUAUUUCCGGCACCGCAAGU ..(((((((((....)))))))))((.((((((((((......)))))...(((........))).))))).))....((((.((....)).)))) ( -37.70) >DroEre_CAF1 8343 93 - 1 GUUGUGGCGGUUUUGGCA---CCAUACGCCACGGGGGAGCUUGCCCCCAACGCCACGGGAUCGGCAGUGGUUGACAUUAUUUCCGGCACCGCAAGU ...((((((....((((.---......)))).(((((......)))))..))))))((..((((.((((((....)))))).))))..))...... ( -38.90) >DroYak_CAF1 143 96 - 1 GUUGUGGUGGUUUUGGCCACUACAUCCGCCACGGGGGAGCUUACCCCCAACGCCACGGGUUCCGCUGUGGUUGACAUUAUUUCCGGCACCGCAAGU ..(((((((((....)))))))))...(((..(((((......)))))...(((((((......))))))).............)))..(....). ( -39.80) >consensus GUUGUGGUGGUUUUGGCCACUACAUCCACCACGGGGGAGUUUACCCCCAGCGCCACGGGAUCGGCAGUGGUUGACAUUAUUUCCGGCACCGCAAGU ..(((((((((....)))))))))((.((((((((((......)))))...(((........))).))))).))...................... (-21.92 = -25.00 + 3.08)

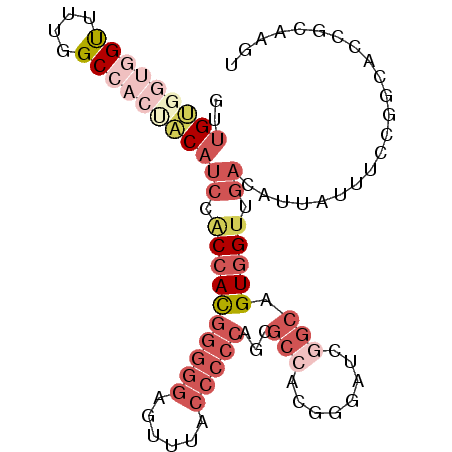
| Location | 21,733,404 – 21,733,497 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 78.11 |
| Mean single sequence MFE | -33.86 |
| Consensus MFE | -20.45 |
| Energy contribution | -22.73 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
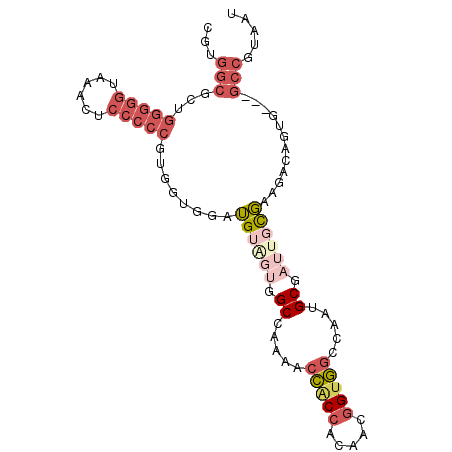
>3R_DroMel_CAF1 21733404 93 + 27905053 CGUGGCGCUGGGGGUAAACUCCCCCGUGGAGGAUGUAGUGGCCAAAACCACCACAACGGUGGCCAAUGCGAUUGCGAAGACAGUG---GCCGUAAU ...(((((((((((......)))))........((((((.((.....(((((.....))))).....)).)))))).....))).---)))..... ( -33.70) >DroGri_CAF1 12809 81 + 1 GG------------UGGCCACAACAAUGGUGGCGGA---CGCUAAUGCUCCAUCUAUAGUAACCAAUGCGAUUGUUCAGACAGUUCAGUCCAUAAU ((------------(.(((((.......)))))(((---.((....)))))..........))).(((.(((((..(.....)..))))))))... ( -23.20) >DroSec_CAF1 25 93 + 1 CGUGGCGCAGGGGGUAAACUCCCCCGUGGUGGAUGUGGUGGCCAAAACCACCACAACGGUGGCCAAUGCGAUUGCGAAGACAGUG---GCCGUAAU ((..((((((((((......))))).((((.(.((((((((......))))))))....).)))).)))).)..)).........---........ ( -39.40) >DroSim_CAF1 182 93 + 1 CGUGGCGCAGGGGGUAAACUCCCCCGUGGUGGAUGUAGUGGCCAAAACCACCACAACGGUGGCCAAUGCGAUUGCGAAGACAGUG---GCCGUAAU ..((((.(((((((......)))))(((((((...............))))))).....)))))).((((.(..(.......)..---).)))).. ( -34.96) >DroEre_CAF1 8382 90 + 1 CGUGGCGUUGGGGGCAAGCUCCCCCGUGGCGUAUGG---UGCCAAAACCGCCACAACGGUGGCCAAUGCGAUUGCGAAGACAGUG---GCCGUAAU .((((((..(((((......))))).(((((.....---)))))....)))))).(((((.((.....((....))......)).---)))))... ( -37.10) >DroYak_CAF1 182 93 + 1 CGUGGCGUUGGGGGUAAGCUCCCCCGUGGCGGAUGUAGUGGCCAAAACCACCACAACGGUGGCCAAUGCGAUUGCGAAGACAGUG---GCCGUAAU ...(((...(((((......)))))...((((.((((.(((((...(((........)))))))).)))).))))..........---)))..... ( -34.80) >consensus CGUGGCGCUGGGGGUAAACUCCCCCGUGGUGGAUGUAGUGGCCAAAACCACCACAACGGUGGCCAAUGCGAUUGCGAAGACAGUG___GCCGUAAU ...(((...(((((......)))))........((((((.((.....(((((.....))))).....)).))))))............)))..... (-20.45 = -22.73 + 2.28)


| Location | 21,733,404 – 21,733,497 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 78.11 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -19.64 |
| Energy contribution | -21.08 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
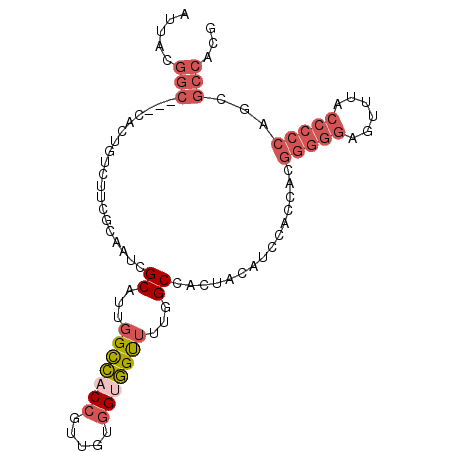
>3R_DroMel_CAF1 21733404 93 - 27905053 AUUACGGC---CACUGUCUUCGCAAUCGCAUUGGCCACCGUUGUGGUGGUUUUGGCCACUACAUCCUCCACGGGGGAGUUUACCCCCAGCGCCACG .....(((---((.(((..........))).)))))..((((((((((((....)))))))).........(((((......)))))))))..... ( -35.70) >DroGri_CAF1 12809 81 - 1 AUUAUGGACUGAACUGUCUGAACAAUCGCAUUGGUUACUAUAGAUGGAGCAUUAGCG---UCCGCCACCAUUGUUGUGGCCA------------CC .....((((....(((((((...((((.....))))....))))))).((....)))---)))(((((.......)))))..------------.. ( -20.20) >DroSec_CAF1 25 93 - 1 AUUACGGC---CACUGUCUUCGCAAUCGCAUUGGCCACCGUUGUGGUGGUUUUGGCCACCACAUCCACCACGGGGGAGUUUACCCCCUGCGCCACG .....(((---((.(((..........))).)))))..((((((((((((....)))))))))........(((((......))))).)))..... ( -36.90) >DroSim_CAF1 182 93 - 1 AUUACGGC---CACUGUCUUCGCAAUCGCAUUGGCCACCGUUGUGGUGGUUUUGGCCACUACAUCCACCACGGGGGAGUUUACCCCCUGCGCCACG .....(((---((.(((..........))).)))))..(((.(((((((...((.......)).)))))))(((((......))))).)))..... ( -35.30) >DroEre_CAF1 8382 90 - 1 AUUACGGC---CACUGUCUUCGCAAUCGCAUUGGCCACCGUUGUGGCGGUUUUGGCA---CCAUACGCCACGGGGGAGCUUGCCCCCAACGCCACG .....(((---((.(((..........))).)))))......((((((....((((.---......)))).(((((......)))))..)))))). ( -38.60) >DroYak_CAF1 182 93 - 1 AUUACGGC---CACUGUCUUCGCAAUCGCAUUGGCCACCGUUGUGGUGGUUUUGGCCACUACAUCCGCCACGGGGGAGCUUACCCCCAACGCCACG .....(((---((.(((..........))).)))))..(((((((((((...((.......)).)))))))(((((......)))))))))..... ( -36.70) >consensus AUUACGGC___CACUGUCUUCGCAAUCGCAUUGGCCACCGUUGUGGUGGUUUUGGCCACUACAUCCACCACGGGGGAGUUUACCCCCAGCGCCACG .....(((...................((...(((((((.....)))))))...))...............(((((......)))))...)))... (-19.64 = -21.08 + 1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:26 2006