
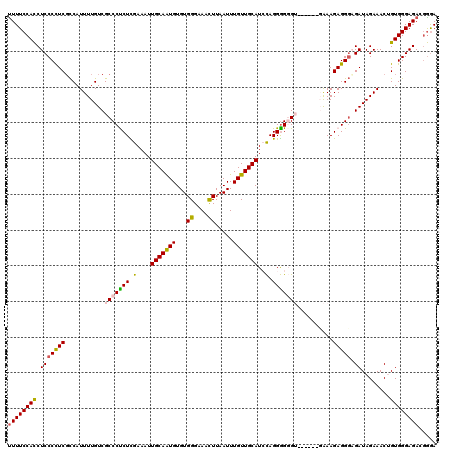
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,709,103 – 21,709,216 |
| Length | 113 |
| Max. P | 0.739050 |

| Location | 21,709,103 – 21,709,216 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.94 |
| Mean single sequence MFE | -40.08 |
| Consensus MFE | -27.74 |
| Energy contribution | -27.74 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21709103 113 + 27905053 UUUUCCAUCUCCCCUCUCCAUUUUGUCGCCCUCUUGAAAUUGCAAUGUGUGGGAAACUUAAUUUGUUGCAUCUAGGAG-GU------GAAAGAGGGAGAUAGAAACUGUGGGAGACGAGA .((((..((((((..((((.((((.(((((.(((((....(((((((...((....)).....)))))))..))))))-))------)).))))))))((((...)))))))))).)))) ( -36.70) >DroSec_CAF1 36490 119 + 1 UUUUCCACCUCCCCUCGCCAUUUUGUCGCCCCCUCGAAAUUGCAAUGUGUGGGAAACUUAAUUUGUUGCAUACAGGGGGGUGG-GGGCAAAGAGGGUGAUAGAAACUGUGGGAGACGGGA ..((((..(((((..((((.(((((((.(((((((.....(((((((...((....)).....))))))).....))))).))-.)))))))..))))((((...)))))))))..)))) ( -46.30) >DroSim_CAF1 48248 120 + 1 UUUUCCACCUCCCCUCGCCAUUUUGUCGCCCCCUUGAAAUUGCAAUGUGUGGGAAACUUAAUUUGUUGCAUACAGGGGGGUGGGGGGCAAAGAGGGUGAUAGAAACUGUGGGAGACGGGA ((((((((.((((((((((....(.(((((((((((....(((((((...((....)).....)))))))..))))))))))).))))...))))).))........))))))))..... ( -47.60) >DroEre_CAF1 33110 112 + 1 UUUUCCACCUCCCUUCGUCAUUUUGUCGCCCUCUCGAAAUUGCAGUGUGUGGGAAACUUAAUUUGUUGCAUCCAGGGG-G-------GAAAGAGGCAGAUAGAAACAGUGGGAGAAUGGA ...((((.(((((....((..((((((.(((((..(....((((..(...((....)).....)..))))..)..)))-)-------).....))))))..))......)))))..)))) ( -34.50) >DroYak_CAF1 33333 113 + 1 CUUUCCACCUCCCCUCGCCAUUUUGUCGCCCUCUCGAAAUUGCAAUGUGUGGGAAAUUUAAUUUGUUGCAUCCAGGGGGG-------GGAAGAGGGAGAUAGAAACAGUGGGAGACGGAA ..((((..(((((((.(...(((((((.((((((......(((((((.((..........)).)))))))(((.......-------))))))))).))))))).))).)))))..)))) ( -35.30) >consensus UUUUCCACCUCCCCUCGCCAUUUUGUCGCCCUCUCGAAAUUGCAAUGUGUGGGAAACUUAAUUUGUUGCAUCCAGGGGGGU______GAAAGAGGGAGAUAGAAACUGUGGGAGACGGGA ((((((((.(((((((...........(((((((.(....(((((((...((....)).....)))))))..).)))))))..........))))).))........))))))))..... (-27.74 = -27.74 + 0.00)


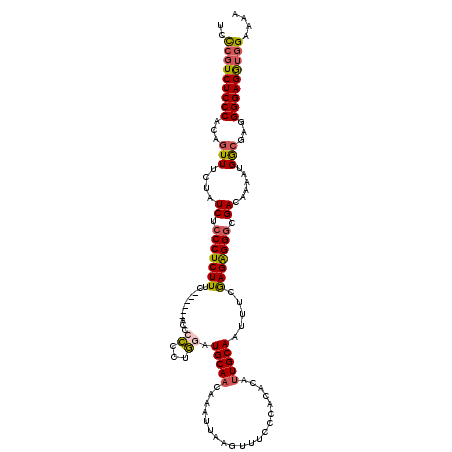
| Location | 21,709,103 – 21,709,216 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.94 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -23.65 |
| Energy contribution | -23.77 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.739050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21709103 113 - 27905053 UCUCGUCUCCCACAGUUUCUAUCUCCCUCUUUC------AC-CUCCUAGAUGCAACAAAUUAAGUUUCCCACACAUUGCAAUUUCAAGAGGGCGACAAAAUGGAGAGGGGAGAUGGAAAA ...............((((((((((((.(((((------.(-(((..((((((((....................)))).))))...))))....(.....)))))))))))))))))). ( -30.55) >DroSec_CAF1 36490 119 - 1 UCCCGUCUCCCACAGUUUCUAUCACCCUCUUUGCCC-CCACCCCCCUGUAUGCAACAAAUUAAGUUUCCCACACAUUGCAAUUUCGAGGGGGCGACAAAAUGGCGAGGGGAGGUGGAAAA ...............((((((((.(((((.((((((-((((......)).(((((....................))))).......))))))))(.....)..)))))..)))))))). ( -32.75) >DroSim_CAF1 48248 120 - 1 UCCCGUCUCCCACAGUUUCUAUCACCCUCUUUGCCCCCCACCCCCCUGUAUGCAACAAAUUAAGUUUCCCACACAUUGCAAUUUCAAGGGGGCGACAAAAUGGCGAGGGGAGGUGGAAAA .....................((((((((((((((...(.(((((.((..(((((....................)))))....)).))))).).......))))))))).))))).... ( -33.35) >DroEre_CAF1 33110 112 - 1 UCCAUUCUCCCACUGUUUCUAUCUGCCUCUUUC-------C-CCCCUGGAUGCAACAAAUUAAGUUUCCCACACACUGCAAUUUCGAGAGGGCGACAAAAUGACGAAGGGAGGUGGAAAA (((((.(((((..((((......((((.(((((-------(-(....)).((((......................)))).....))))))))).......))))..))))))))))... ( -29.07) >DroYak_CAF1 33333 113 - 1 UUCCGUCUCCCACUGUUUCUAUCUCCCUCUUCC-------CCCCCCUGGAUGCAACAAAUUAAAUUUCCCACACAUUGCAAUUUCGAGAGGGCGACAAAAUGGCGAGGGGAGGUGGAAAG ((((..(((((..((((....((.(((((((.(-------(......)).(((((....................))))).....))))))).))......))))..)))))..)))).. ( -33.95) >consensus UCCCGUCUCCCACAGUUUCUAUCUCCCUCUUUC______ACCCCCCUGGAUGCAACAAAUUAAGUUUCCCACACAUUGCAAUUUCGAGAGGGCGACAAAAUGGCGAGGGGAGGUGGAAAA ..(((((((((...(((....((.(((((((...........((...)).(((((....................))))).....))))))).))......)))...))))))))).... (-23.65 = -23.77 + 0.12)

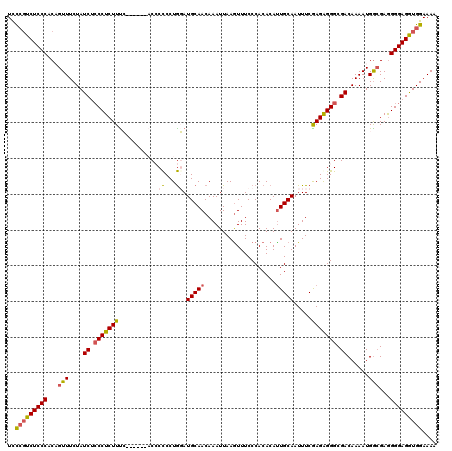
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:14 2006