
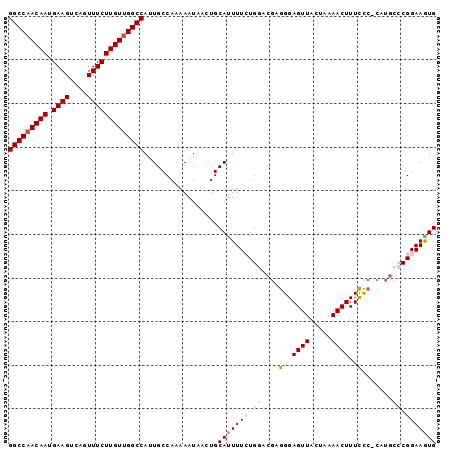
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,686,981 – 21,687,117 |
| Length | 136 |
| Max. P | 0.990963 |

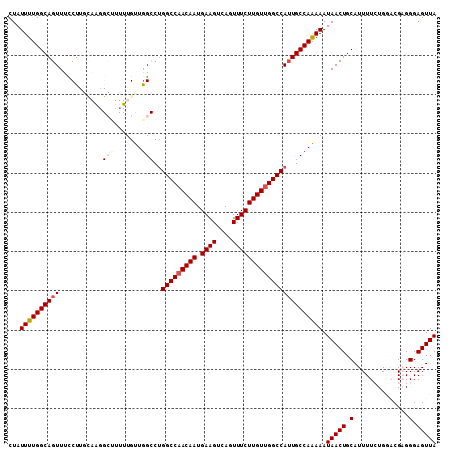
| Location | 21,686,981 – 21,687,090 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 88.15 |
| Mean single sequence MFE | -35.03 |
| Consensus MFE | -27.80 |
| Energy contribution | -28.57 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21686981 109 + 27905053 CUAUUUUGGCAGUUUCCUUACAAGGCUUUUUGUUGGCCUGGCCAACAAUGAAGUCAGUUUCUUGUUGGCCAUUGCCAAAAAUAACUGCAUUUUCUGGACGAGGGAGUUA ...((((((((((.........(((((.......)))))(((((((((.((((....)))))))))))))))))))))))........(((..((.....))..))).. ( -37.30) >DroPse_CAF1 76950 94 + 1 CUAUUUUGGCAGUUUUCU------------UGUUGG-CUGGCCAACAAUGAAGCCAGUUUCUUGUUGGCCAUUGCCAGAAAUAACUGCAUUUUGUG--CGCGGGAGUUA ..(((((.((((((....------------..((((-(((((((((((.((((....))))))))))))))..)))))....))))((((...)))--))).))))).. ( -34.90) >DroEre_CAF1 10865 109 + 1 CUAUUUUGGCAGUUUCCUUGCAGGGCUUUUUGUUGGCCUGGCCAACAAUGAAGUCAGUUUCUUGUUGGCCAAUGCCAAAAAUAACUGCAUUUUCUGGACGAGGGAGUUA ((.(((((.(((......(((((.........(((((.((((((((((.((((....))))))))))))))..)))))......)))))....)))..))))).))... ( -36.16) >DroYak_CAF1 10996 109 + 1 CUAUUUUGGCAGUUUCCUUGCAAGGCUUUUUGUUAGCCUGGCCAACAAUGAAGUCAGUUUCUUGUUGGCCAUUGCCAAAAAUAACUGCAUUUUCUGGACGAGGGAGUUA ...((((((((((.........(((((.......)))))(((((((((.((((....)))))))))))))))))))))))........(((..((.....))..))).. ( -37.30) >DroAna_CAF1 12063 109 + 1 CUAUUUUGGCAGUUUCCUUGUAACGCUUUUUGUUGGCCCGGCCAACAAUGAAGUCAGUUUCUUGUCGGCCAUUGCCAAAAAUAACUGCAUUUUCUGGACGAGGGAGUUA ............(((((((((...(((((((((((((...)))))))).)))))(((((..((...(((....)))..))..)))))..........)))))))))... ( -29.60) >DroPer_CAF1 76706 94 + 1 CUAUUUUGGCAGUUUUCU------------UGUUGG-CUGGCCAACAAUGAAGCCAGUUUCUUGUUGGCCAUUGCCAGAAAUAACUGCAUUUUGUG--CGCGGGAGUUA ..(((((.((((((....------------..((((-(((((((((((.((((....))))))))))))))..)))))....))))((((...)))--))).))))).. ( -34.90) >consensus CUAUUUUGGCAGUUUCCUUGCAAGGCUUUUUGUUGGCCUGGCCAACAAUGAAGUCAGUUUCUUGUUGGCCAUUGCCAAAAAUAACUGCAUUUUCUGGACGAGGGAGUUA ...((((((((((...........(((.......)))..(((((((((.((((....))))))))))))))))))))))).(((((.(..............).))))) (-27.80 = -28.57 + 0.78)


| Location | 21,686,981 – 21,687,090 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 88.15 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -26.53 |
| Energy contribution | -26.58 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.94 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21686981 109 - 27905053 UAACUCCCUCGUCCAGAAAAUGCAGUUAUUUUUGGCAAUGGCCAACAAGAAACUGACUUCAUUGUUGGCCAGGCCAACAAAAAGCCUUGUAAGGAAACUGCCAAAAUAG .....................((((((....(((((..(((((((((((((......))).)))))))))).))))).......(((....))).))))))........ ( -33.20) >DroPse_CAF1 76950 94 - 1 UAACUCCCGCG--CACAAAAUGCAGUUAUUUCUGGCAAUGGCCAACAAGAAACUGGCUUCAUUGUUGGCCAG-CCAACA------------AGAAAACUGCCAAAAUAG ...........--........((((((.(((.((((..(((((((((((((......))).)))))))))))-)))...------------))).))))))........ ( -30.10) >DroEre_CAF1 10865 109 - 1 UAACUCCCUCGUCCAGAAAAUGCAGUUAUUUUUGGCAUUGGCCAACAAGAAACUGACUUCAUUGUUGGCCAGGCCAACAAAAAGCCCUGCAAGGAAACUGCCAAAAUAG ....................(((((...((((((((.((((((((((((((......))).)))))))))))))))...))))...)))))((....)).......... ( -34.30) >DroYak_CAF1 10996 109 - 1 UAACUCCCUCGUCCAGAAAAUGCAGUUAUUUUUGGCAAUGGCCAACAAGAAACUGACUUCAUUGUUGGCCAGGCUAACAAAAAGCCUUGCAAGGAAACUGCCAAAAUAG .....................((((((.((((((.((((((((((((((((......))).))))))))))((((.......)))))))))))))))))))........ ( -34.60) >DroAna_CAF1 12063 109 - 1 UAACUCCCUCGUCCAGAAAAUGCAGUUAUUUUUGGCAAUGGCCGACAAGAAACUGACUUCAUUGUUGGCCGGGCCAACAAAAAGCGUUACAAGGAAACUGCCAAAAUAG ...........(((....(((((..((....(((((..(((((((((((((......))).)))))))))).)))))..))..)))))....))).............. ( -29.20) >DroPer_CAF1 76706 94 - 1 UAACUCCCGCG--CACAAAAUGCAGUUAUUUCUGGCAAUGGCCAACAAGAAACUGGCUUCAUUGUUGGCCAG-CCAACA------------AGAAAACUGCCAAAAUAG ...........--........((((((.(((.((((..(((((((((((((......))).)))))))))))-)))...------------))).))))))........ ( -30.10) >consensus UAACUCCCUCGUCCAGAAAAUGCAGUUAUUUUUGGCAAUGGCCAACAAGAAACUGACUUCAUUGUUGGCCAGGCCAACAAAAAGCCUUGCAAGGAAACUGCCAAAAUAG .............................(((((((..(((((((((((((......))).))))))))))....................((....)))))))))... (-26.53 = -26.58 + 0.06)



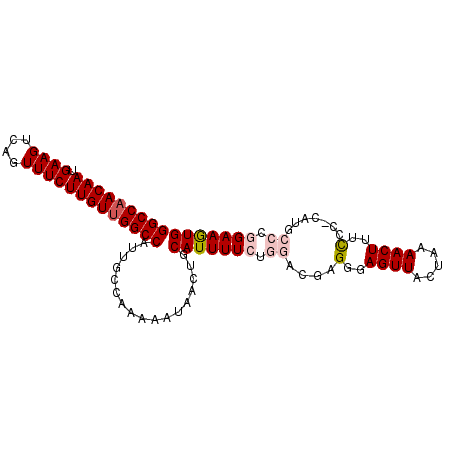
| Location | 21,687,020 – 21,687,117 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 89.78 |
| Mean single sequence MFE | -32.06 |
| Consensus MFE | -24.13 |
| Energy contribution | -24.93 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21687020 97 + 27905053 GGCCAACAAUGAAGUCAGUUUCUUGUUGGCCAUUGCCAAAAAUAACUGCAUUUUCUGGACGAGGGAGUUAGUAAAACUUUCCC-CAUACCCGGAAGUG (((((((((.((((....)))))))))))))....................(((((((....(((.(..((.....))..)))-)....))))))).. ( -29.80) >DroPse_CAF1 76976 95 + 1 GGCCAACAAUGAAGCCAGUUUCUUGUUGGCCAUUGCCAGAAAUAACUGCAUUUUGUG--CGCGGGAGUUACGAAAACUUUCUC-UCUGCACGGAAAUG (((((((((.((((....)))))))))))))..(((.((......)))))(((((((--((.(((((.............)))-)))))))))))... ( -34.82) >DroSec_CAF1 10009 96 + 1 GGCCAACAAUGAAGUCAGUUUCUUGUUGGCCAUUGCCAA-AAUAACUGCAUUUUCUGGACGAGGGAGUUAGUAAAACUUUCCC-CAUGCCCGGAAGUG (((((((((.((((....)))))))))))))..(((...-.......))).(((((((.((.(((.(..((.....))..)))-).)).))))))).. ( -32.10) >DroEre_CAF1 10904 97 + 1 GGCCAACAAUGAAGUCAGUUUCUUGUUGGCCAAUGCCAAAAAUAACUGCAUUUUCUGGACGAGGGAGUUAGUAAAACUUUCUA-CAUGCCCGGAAGUG (((((((((.((((....))))))))))))).................(((((((.((...((..((((.....))))..)).-....)).))))))) ( -31.70) >DroAna_CAF1 12102 98 + 1 GGCCAACAAUGAAGUCAGUUUCUUGUCGGCCAUUGCCAAAAAUAACUGCAUUUUCUGGACGAGGGAGUUACUAAAACUUUUCCCCUGGCCCGGAAGUG ((((.((((.((((....)))))))).))))....................(((((((.(.((((((((.....))))....)))).).))))))).. ( -29.10) >DroPer_CAF1 76732 95 + 1 GGCCAACAAUGAAGCCAGUUUCUUGUUGGCCAUUGCCAGAAAUAACUGCAUUUUGUG--CGCGGGAGUUACGAAAACUUUCUC-UCUGCACGGAAAUG (((((((((.((((....)))))))))))))..(((.((......)))))(((((((--((.(((((.............)))-)))))))))))... ( -34.82) >consensus GGCCAACAAUGAAGUCAGUUUCUUGUUGGCCAUUGCCAAAAAUAACUGCAUUUUCUGGACGAGGGAGUUACUAAAACUUUCCC_CAUGCCCGGAAGUG (((((((((.((((....))))))))))))).................(((((((.((....(..((((.....))))..).......)).))))))) (-24.13 = -24.93 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:03 2006