
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,685,250 – 21,685,340 |
| Length | 90 |
| Max. P | 0.711214 |

| Location | 21,685,250 – 21,685,340 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 73.26 |
| Mean single sequence MFE | -17.65 |
| Consensus MFE | -5.06 |
| Energy contribution | -5.39 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
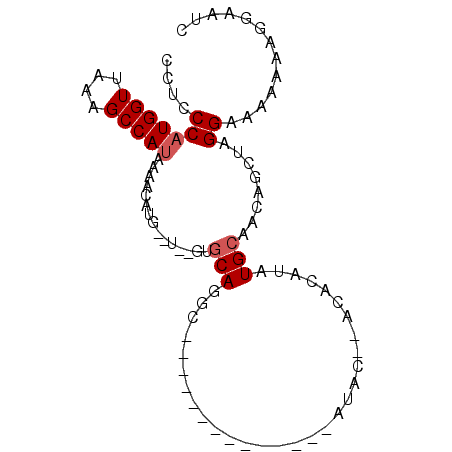
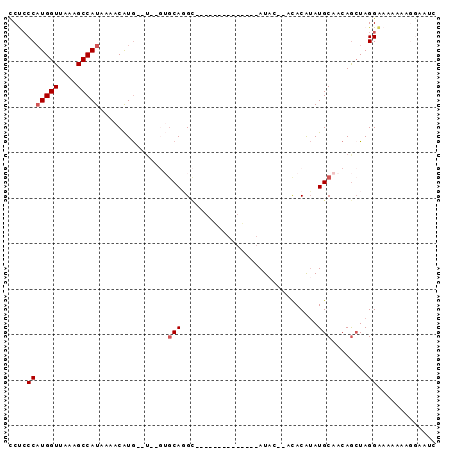
>3R_DroMel_CAF1 21685250 90 + 27905053 CCUCCCAUGGUUAAAGCCAUAAAACAUGCGU--GUGCAGGCAUCGGGUGUGCUACAUAC--ACACAUAUGCAACAGCUAGGAAAAAAAGGAAUC ..(((.(((((....))))).......((.(--((....((((...(((((.......)--))))..)))).)))))..)))............ ( -20.90) >DroSec_CAF1 8330 74 + 1 CCUCCCAUGGUUAAAGCCAUAAAACAUG--U--GUGCAGGC--------------AUAC--ACACAUAUGCAACAGCUAGGAAAAAAAGGAAUC ..(((.(((((....))))).....(((--(--(((.....--------------...)--))))))..((....))..)))............ ( -14.90) >DroSim_CAF1 8730 74 + 1 CCUCCCAUGGUUAAAGCCAUAAAACAUG--U--GUGCAGGC--------------AUAC--ACACAUAUGCAACAGCUAGGAAAAAAAGGAAUC ..(((.(((((....))))).....(((--(--(((.....--------------...)--))))))..((....))..)))............ ( -14.90) >DroEre_CAF1 9090 85 + 1 UCU-CCAUGGUUAAAGCCAUAAAACAUG--UGUGUGCAGGCAUCGCAUGUGGCACAU----ACACAUAUGCAACAGCUAGGAA--AACCGAAUC ..(-(((((((....)))))......((--(((((((..((((...)))).))))))----))).....((....))..))).--......... ( -22.70) >DroAna_CAF1 10131 81 + 1 GCCCCCAUGGUUAAAGCCAUAAAGCGUG--U----GCAUGU-----GUAUAAAAAAUACGUAUACAUAUGCAACAAAUAGGGAAAAA--UAAAC ...((((((((....))))).....((.--(----((((((-----(((((.........)))))))))))))).....))).....--..... ( -24.10) >DroPer_CAF1 74540 69 + 1 UCCCCCCUGGUUAAGGCCAUAAAACAUA--------CAUGG--------------AUAC--ACACGAAUGCAACAGCUAGGAAA-AAAGUAAAC .....(((((((..(.((((........--------.))))--------------...)--....(....)...)))))))...-......... ( -8.40) >consensus CCUCCCAUGGUUAAAGCCAUAAAACAUG__U__GUGCAGGC______________AUAC__ACACAUAUGCAACAGCUAGGAAAAAAAGGAAUC ....(((((((....)))))...............(((..............................)))........))............. ( -5.06 = -5.39 + 0.33)

| Location | 21,685,250 – 21,685,340 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 73.26 |
| Mean single sequence MFE | -21.51 |
| Consensus MFE | -6.68 |
| Energy contribution | -6.68 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
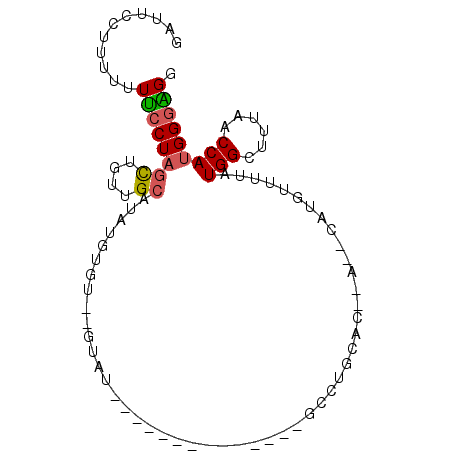
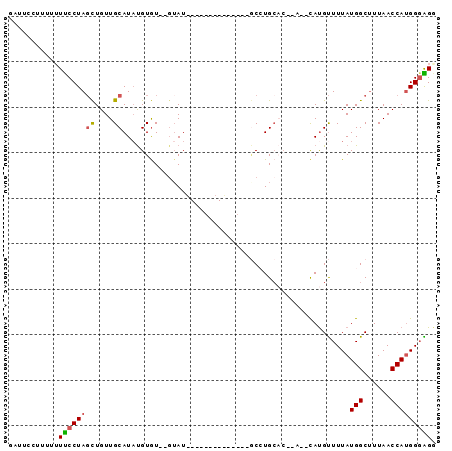
>3R_DroMel_CAF1 21685250 90 - 27905053 GAUUCCUUUUUUUCCUAGCUGUUGCAUAUGUGU--GUAUGUAGCACACCCGAUGCCUGCAC--ACGCAUGUUUUAUGGCUUUAACCAUGGGAGG ..(((((.........((((((.((((.(((((--(((.(((.(......).))).)))))--))).))))...))))))........))))). ( -26.33) >DroSec_CAF1 8330 74 - 1 GAUUCCUUUUUUUCCUAGCUGUUGCAUAUGUGU--GUAU--------------GCCUGCAC--A--CAUGUUUUAUGGCUUUAACCAUGGGAGG ..(((((.........((((((...((((((((--(((.--------------...)))))--)--)))))...))))))........))))). ( -23.83) >DroSim_CAF1 8730 74 - 1 GAUUCCUUUUUUUCCUAGCUGUUGCAUAUGUGU--GUAU--------------GCCUGCAC--A--CAUGUUUUAUGGCUUUAACCAUGGGAGG ..(((((.........((((((...((((((((--(((.--------------...)))))--)--)))))...))))))........))))). ( -23.83) >DroEre_CAF1 9090 85 - 1 GAUUCGGUU--UUCCUAGCUGUUGCAUAUGUGU----AUGUGCCACAUGCGAUGCCUGCACACA--CAUGUUUUAUGGCUUUAACCAUGG-AGA .....((((--.....((((((...((((((((----..((((..(((...)))...)))))))--)))))...))))))..))))....-... ( -21.90) >DroAna_CAF1 10131 81 - 1 GUUUA--UUUUUCCCUAUUUGUUGCAUAUGUAUACGUAUUUUUUAUAC-----ACAUGC----A--CACGCUUUAUGGCUUUAACCAUGGGGGC .....--....(((((((..(((((((.((((((.........)))))-----).))))----)--.))(((....))).......))))))). ( -19.80) >DroPer_CAF1 74540 69 - 1 GUUUACUUU-UUUCCUAGCUGUUGCAUUCGUGU--GUAU--------------CCAUG--------UAUGUUUUAUGGCCUUAACCAGGGGGGA .......((-(..(((.(((((.((((.((((.--....--------------.))))--------.))))...))))).......)))..))) ( -13.40) >consensus GAUUCCUUUUUUUCCUAGCUGUUGCAUAUGUGU__GUAU______________GCCUGCAC__A__CAUGUUUUAUGGCUUUAACCAUGGGAGG ...........((((((((....))..................................................(((......))))))))). ( -6.68 = -6.68 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:02:00 2006