
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,675,206 – 21,675,344 |
| Length | 138 |
| Max. P | 0.999740 |

| Location | 21,675,206 – 21,675,311 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 94.86 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -26.14 |
| Energy contribution | -26.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21675206 105 + 27905053 ACUGUGACAAAUAUACGGUGUAAAUCAUUGAGCUGCCUGCUUCUACAGGCUGCUCCACACGUGUUAAAUUUAACGCAAGAAAAGGUGCAUUAAAUAUGCAAGAGU ..((((..........(((....)))...((((.(((((......))))).)))))))).((((((....)))))).........(((((.....)))))..... ( -28.30) >DroSec_CAF1 155193 105 + 1 ACUGUGACAAAUAUGCGGUGUAAAUCAUUGAGCUGCCUGCUUCUACAGGCUGCCCCACACGUGUUAAAUUUAACGCAAGAAAAGGUGCAUUAAAUAUGCAAGAGU ..((((.........(((((.....))))).((.(((((......))))).))..)))).((((((....)))))).........(((((.....)))))..... ( -25.90) >DroSim_CAF1 160469 105 + 1 ACUGUGACAAAUAUGCGGUGUAAAUCAUUGAGCUGCCUGCUUCUACAGGCUGCCCCACACGUGUUAAAUUUAACGCAAGAAAAGGUGCAUUAAAUAUGCAAGAUU ..((((.........(((((.....))))).((.(((((......))))).))..)))).((((((....)))))).........(((((.....)))))..... ( -25.90) >DroEre_CAF1 152253 105 + 1 ACUGUGACAAAUAUAUGGUGUAAAUCAUUGAGCUGCCUGCUUCUACAGGCAGCUCCACACGUGUUGAAUUUAACGCAGGAAAAGGUGCAUUAAAUAUGCAGGAGU .((((.....((((.(((((((.......((((((((((......)))))))))).....((((((....)))))).........))))))).)))))))).... ( -33.90) >DroYak_CAF1 153268 105 + 1 ACUGUGACAAAUAUACGGUGUAAAUCAUUGAGCUGCCUGCCUCUACAGGCAGCUCCACACGUGUUGAAUUUGACGCAAGAAAAGGUGCAUUAAAUAUGCAGGCGU ..((((.(((((((((((((.........((((((((((......))))))))))))).)))))...))))).))))........(((((.....)))))..... ( -35.50) >consensus ACUGUGACAAAUAUACGGUGUAAAUCAUUGAGCUGCCUGCUUCUACAGGCUGCUCCACACGUGUUAAAUUUAACGCAAGAAAAGGUGCAUUAAAUAUGCAAGAGU ..((((..........(((....)))...((((.(((((......))))).)))))))).((((((....)))))).........(((((.....)))))..... (-26.14 = -26.14 + 0.00)



| Location | 21,675,206 – 21,675,311 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 94.86 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -26.95 |
| Energy contribution | -26.87 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.98 |
| SVM RNA-class probability | 0.999740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21675206 105 - 27905053 ACUCUUGCAUAUUUAAUGCACCUUUUCUUGCGUUAAAUUUAACACGUGUGGAGCAGCCUGUAGAAGCAGGCAGCUCAAUGAUUUACACCGUAUAUUUGUCACAGU (((.(.(((.((((((((((........)))))))))).....(((.(((((((.((((((....)))))).))))((....)).)))))).....))).).))) ( -28.30) >DroSec_CAF1 155193 105 - 1 ACUCUUGCAUAUUUAAUGCACCUUUUCUUGCGUUAAAUUUAACACGUGUGGGGCAGCCUGUAGAAGCAGGCAGCUCAAUGAUUUACACCGCAUAUUUGUCACAGU .....(((..((((((((((........)))))))))).......(((((((((.((((((....)))))).)))).......))))).)))............. ( -28.71) >DroSim_CAF1 160469 105 - 1 AAUCUUGCAUAUUUAAUGCACCUUUUCUUGCGUUAAAUUUAACACGUGUGGGGCAGCCUGUAGAAGCAGGCAGCUCAAUGAUUUACACCGCAUAUUUGUCACAGU .....(((..((((((((((........)))))))))).......(((((((((.((((((....)))))).)))).......))))).)))............. ( -28.71) >DroEre_CAF1 152253 105 - 1 ACUCCUGCAUAUUUAAUGCACCUUUUCCUGCGUUAAAUUCAACACGUGUGGAGCUGCCUGUAGAAGCAGGCAGCUCAAUGAUUUACACCAUAUAUUUGUCACAGU ..........((((((((((........))))))))))........(((((((((((((((....))))))))))).(((........)))........)))).. ( -33.20) >DroYak_CAF1 153268 105 - 1 ACGCCUGCAUAUUUAAUGCACCUUUUCUUGCGUCAAAUUCAACACGUGUGGAGCUGCCUGUAGAGGCAGGCAGCUCAAUGAUUUACACCGUAUAUUUGUCACAGU .....(((((.....))))).......(((.(.(((((.....(((.((((((((((((((....)))))))))))((....)).))))))..))))).).))). ( -33.00) >consensus ACUCUUGCAUAUUUAAUGCACCUUUUCUUGCGUUAAAUUUAACACGUGUGGAGCAGCCUGUAGAAGCAGGCAGCUCAAUGAUUUACACCGUAUAUUUGUCACAGU .....(((..((((((((((........)))))))))).......(((((((((.((((((....)))))).)))).......))))).)))............. (-26.95 = -26.87 + -0.08)



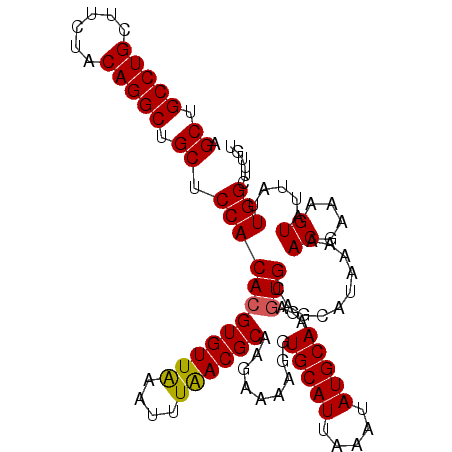
| Location | 21,675,236 – 21,675,344 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -25.50 |
| Energy contribution | -25.30 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21675236 108 + 27905053 AGCUGCCUGCUUCUACAGGCUGCUCCACACGUGUUAAAUUUAACGCAAGAAAAGGUGCAUUAAAUAUGCAAGAGUGCAGCAUAAAAACGAAAGUAUUAUUGGCUUUGU (((((((((......)))))((((.(((..((((((....)))))).........(((((.....)))))...))).)))).....((....))......)))).... ( -30.40) >DroSec_CAF1 155223 108 + 1 AGCUGCCUGCUUCUACAGGCUGCCCCACACGUGUUAAAUUUAACGCAAGAAAAGGUGCAUUAAAUAUGCAAGAGUGCAGCAUAAAAACGAAAGUAUUAUUGGCUUUGU (((((((((......)))))(((..(((..((((((....)))))).........(((((.....)))))...)))..))).....((....))......)))).... ( -28.40) >DroSim_CAF1 160499 108 + 1 AGCUGCCUGCUUCUACAGGCUGCCCCACACGUGUUAAAUUUAACGCAAGAAAAGGUGCAUUAAAUAUGCAAGAUUGCAGCAUAAAAACGAAAGUAUUAUUGGCUUUGU ....(((...........(((((.......((((((....)))))).........(((((.....))))).....)))))......((....))......)))..... ( -25.10) >DroEre_CAF1 152283 108 + 1 AGCUGCCUGCUUCUACAGGCAGCUCCACACGUGUUGAAUUUAACGCAGGAAAAGGUGCAUUAAAUAUGCAGGAGUGCAGGAUAAAAACGAAAGUAUUAUUGGCUUUGU .((((((((......))))))))(((....((((((....)))))).........(((((.....))))))))..((..(((((..((....)).))))).))..... ( -33.60) >DroYak_CAF1 153298 108 + 1 AGCUGCCUGCCUCUACAGGCAGCUCCACACGUGUUGAAUUUGACGCAAGAAAAGGUGCAUUAAAUAUGCAGGCGUGCAGGAUAAAAACGAAAGUUUUAUUGGCUUUGU .((((((((......))))))))(((.(((((...........(....)......(((((.....))))).)))))..)))..(((((....)))))........... ( -35.20) >consensus AGCUGCCUGCUUCUACAGGCUGCUCCACACGUGUUAAAUUUAACGCAAGAAAAGGUGCAUUAAAUAUGCAAGAGUGCAGCAUAAAAACGAAAGUAUUAUUGGCUUUGU .((.(((((......))))).)).((((((((((((....)))))).........(((((.....)))))...)))..........((....)).....)))...... (-25.50 = -25.30 + -0.20)


| Location | 21,675,236 – 21,675,344 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -31.66 |
| Consensus MFE | -27.04 |
| Energy contribution | -28.00 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21675236 108 - 27905053 ACAAAGCCAAUAAUACUUUCGUUUUUAUGCUGCACUCUUGCAUAUUUAAUGCACCUUUUCUUGCGUUAAAUUUAACACGUGUGGAGCAGCCUGUAGAAGCAGGCAGCU ....(((....................((((.(((.(.((...((((((((((........))))))))))....)).).))).))))((((((....)))))).))) ( -32.00) >DroSec_CAF1 155223 108 - 1 ACAAAGCCAAUAAUACUUUCGUUUUUAUGCUGCACUCUUGCAUAUUUAAUGCACCUUUUCUUGCGUUAAAUUUAACACGUGUGGGGCAGCCUGUAGAAGCAGGCAGCU ....(((....................((((.(((.(.((...((((((((((........))))))))))....)).).))).))))((((((....)))))).))) ( -32.00) >DroSim_CAF1 160499 108 - 1 ACAAAGCCAAUAAUACUUUCGUUUUUAUGCUGCAAUCUUGCAUAUUUAAUGCACCUUUUCUUGCGUUAAAUUUAACACGUGUGGGGCAGCCUGUAGAAGCAGGCAGCU .....(((....((((.........(((((.........)))))(((((((((........)))))))))........))))..))).((((((....)))))).... ( -29.90) >DroEre_CAF1 152283 108 - 1 ACAAAGCCAAUAAUACUUUCGUUUUUAUCCUGCACUCCUGCAUAUUUAAUGCACCUUUUCCUGCGUUAAAUUCAACACGUGUGGAGCUGCCUGUAGAAGCAGGCAGCU ...........................(((.((((........((((((((((........)))))))))).......)))))))(((((((((....))))))))). ( -32.56) >DroYak_CAF1 153298 108 - 1 ACAAAGCCAAUAAAACUUUCGUUUUUAUCCUGCACGCCUGCAUAUUUAAUGCACCUUUUCUUGCGUCAAAUUCAACACGUGUGGAGCUGCCUGUAGAGGCAGGCAGCU ...........(((((....)))))..(((.(((((.......((((.(((((........))))).))))......))))))))(((((((((....))))))))). ( -31.82) >consensus ACAAAGCCAAUAAUACUUUCGUUUUUAUGCUGCACUCUUGCAUAUUUAAUGCACCUUUUCUUGCGUUAAAUUUAACACGUGUGGAGCAGCCUGUAGAAGCAGGCAGCU ....(((....................((((.(((.(.((...((((((((((........))))))))))....)).).))).))))((((((....)))))).))) (-27.04 = -28.00 + 0.96)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:58 2006