
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,736,803 – 2,736,904 |
| Length | 101 |
| Max. P | 0.516317 |

| Location | 2,736,803 – 2,736,904 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.94 |
| Mean single sequence MFE | -17.71 |
| Consensus MFE | -10.78 |
| Energy contribution | -11.03 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2736803 101 + 27905053 UUCCGGUCAAAGUGAUUUAAUUUCCUGGCUCAUUAUCACGAGUCUCUGACCUCAUAAAUCACA--AAAUUGCCCGCU--CCAGCUUCAGUUGGACG------AUAAUCCAG ....((.....((((((((...((..(((((........)))))...)).....)))))))).--........((.(--(((((....))))))))------.....)).. ( -23.90) >DroVir_CAF1 52043 78 + 1 UUCCGGUCAAAGUGAUUUAAUUUCCUGGCUCAUUAUCACGAGUCUCUGACCUCAUAAAUCAAA--A---------CU--CGA--------------------ACAAUGCAG ....(((((.((.((.......))))(((((........)))))..)))))............--.---------..--...--------------------......... ( -12.60) >DroPse_CAF1 32375 110 + 1 UUCCGGUCAAAGUGAUUUAAUUUCCGGGCUCAUUAUCACGAGUCUCUGACCUCAUAAAUCACACAGAAAAGCCCCCUGCCCCCCACCACACAC-ACACACAAACAUUGCAG ....(((....((((((((...((.((((((........))))).).)).....)))))))).(((.........)))......)))......-................. ( -17.00) >DroSec_CAF1 12731 101 + 1 UUCCGGUCAAAGUGAUUUAAUUUCCUGGCUCAUUAUCACGAGUCUCUGAACUCAUAAAUCAUA--AAAUUGCCCGCU--CCAACUCCAGUUGGUCG------AUAAUCCAG ...(((.(((.((((((((..(((..(((((........)))))...)))....)))))))).--...))).)))..--(((((....)))))...------......... ( -21.40) >DroMoj_CAF1 53935 78 + 1 UUCCGGUCAAAGUGAUUUAAUUUCCUGGCUCAUUAUCACGAGUCUCUGACCUCAUAAAUCAAA--A---------CU--CGA--------------------ACAAUGCAG ....(((((.((.((.......))))(((((........)))))..)))))............--.---------..--...--------------------......... ( -12.60) >DroAna_CAF1 11991 107 + 1 UUCCGGUCAAAGUGAUUUAAUUUCCUGGCUCAUUAUCACGAGUCUCUGCCCUCAUAAAUCACA--AAUGCCCCCCCC--CCCGCGCCAGUUCGCCAGCCCGAAUAAUCCAG (((.(((....((((((((.......(((((........)))))..........)))))))).--............--...(((......)))..))).)))........ ( -18.73) >consensus UUCCGGUCAAAGUGAUUUAAUUUCCUGGCUCAUUAUCACGAGUCUCUGACCUCAUAAAUCACA__AAA__GCCC_CU__CCA_C_CCAGUU_G_C_______ACAAUCCAG ...........((((((((.......(((((........)))))..((....))))))))))................................................. (-10.78 = -11.03 + 0.25)

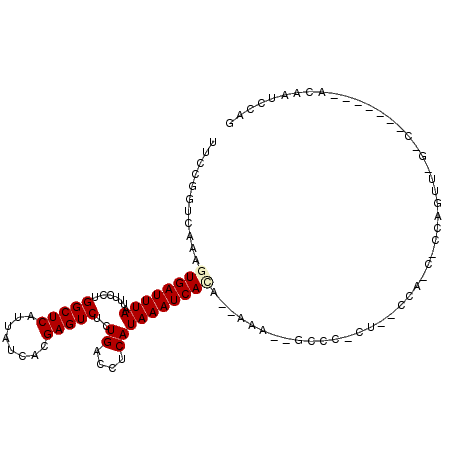
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:41 2006