
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,643,681 – 21,643,803 |
| Length | 122 |
| Max. P | 0.994214 |

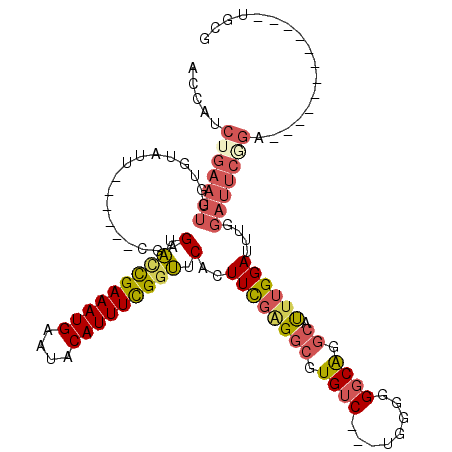
| Location | 21,643,681 – 21,643,780 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.85 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -18.75 |
| Energy contribution | -19.67 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21643681 99 - 27905053 ACCAUCUGAAUUGGUGUAUU------CGAGUACCGAAAUGAAUACAUUUCGGUUCACUUCGAGGCGUGUC---CGGGGGGCAGGCAUUUGGAUUUUGAUUCGGA------------UGCG ..(((((((((..(...(((------((((((((((((((....)))))))))..........((.((((---(...))))).)))))))))).)..)))))))------------)).. ( -39.30) >DroSec_CAF1 132993 99 - 1 ACCAUCUGAAUUGGUGUAUU------CGAGUACCGAAAUGAAUACAUUUCGGUUCACUUCGGGGCGUGUC---UGGGGGGCAGGCAUUUGGAUUUGGAUUCGGU------------UGGG .(((.(((((((.(...(((------((((((((((((((....)))))))))...(....).((.((((---.....)))).)))))))))).).))))))).------------))). ( -37.20) >DroSim_CAF1 129605 99 - 1 ACCAUCUGAAUUGGUGUAUU------CGAGUACCGAAAUGAAUACAUUUCGGUUCACUUCGGGGCGUGUC---UGGGGGGCAGGCAUUUGGAUUUGGAUUCGGU------------UGGG .(((.(((((((.(...(((------((((((((((((((....)))))))))...(....).((.((((---.....)))).)))))))))).).))))))).------------))). ( -37.20) >DroEre_CAF1 129735 111 - 1 ACCAUCUGAAUUGGUGAAUU------CGAGUACCGAAAUGAAUACAUUUCGGUUCAGUUCGAGGCGUGUC---UGUGGGGCAGGCAUUUGGAUUUGGAUUCAGAUUCUGUUUCUGAUUCU ((((.......))))(((((------((((((((((((((....)))))))))..........((.((((---.....)))).))))))))))))(((.(((((.......))))).))) ( -36.30) >DroYak_CAF1 130274 105 - 1 ACCAUCUGAAUUGGUGUAUUCGAGUUCGAGUAUCGAAAUGAAUACAUUUCGCUUCAGUUUGAGGCGUGUC---UGGGUGGCAGGCAUUUGGAUUUGGAUUCCGA------------UUCG .......(((((((...((((((((((((((...((((((....))))))(((((.....)))))..(((---((.....))))))))))))))))))).))))------------))). ( -39.00) >DroAna_CAF1 112525 82 - 1 UCCGUUCGCAUUGG--------------AGUGUUGAAAUGAAUACAUUUUGGCCCAGUUUGUGGCAUGACGAGUGGGGGGCGGUGACUAGGAUUUG------------------------ .((((((...(..(--------------(((((..........))))))..)((((.(((((......))))))))))))))).............------------------------ ( -21.70) >consensus ACCAUCUGAAUUGGUGUAUU______CGAGUACCGAAAUGAAUACAUUUCGGUUCACUUCGAGGCGUGUC___UGGGGGGCAGGCAUUUGGAUUUGGAUUCGGA____________UGCG .....(((((((.................(.(((((((((....))))))))).)..((((((((.((((........)))).)).))))))....)))))))................. (-18.75 = -19.67 + 0.92)

| Location | 21,643,709 – 21,643,803 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.35 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -17.13 |
| Energy contribution | -18.85 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21643709 94 + 27905053 CCCCCCG---GACACGCCUCGAAGUGAACCGAAAUGUAUUCAUUUCGGUACUCG------AAUACACCAAUUCAGAUGGUGCCAGUUUUUGUUAUCCGGGGUC----------------- ..(((((---((......((((.((..(((((((((....))))))))))))))------)...(((((.......))))).............)))))))..----------------- ( -35.10) >DroSec_CAF1 133021 94 + 1 CCCCCCA---GACACGCCCCGAAGUGAACCGAAAUGUAUUCAUUUCGGUACUCG------AAUACACCAAUUCAGAUGGUGCCAGUUUUUGUUAUCCGGGGUC----------------- .......---.....((((((..(.(.(((((((((....))))))))).).).------....(((((.......)))))...............)))))).----------------- ( -28.80) >DroSim_CAF1 129633 94 + 1 CCCCCCA---GACACGCCCCGAAGUGAACCGAAAUGUAUUCAUUUCGGUACUCG------AAUACACCAAUUCAGAUGGUGCCAGUUUUUGUUAUCCGGGGUC----------------- .......---.....((((((..(.(.(((((((((....))))))))).).).------....(((((.......)))))...............)))))).----------------- ( -28.80) >DroEre_CAF1 129775 94 + 1 CCCCACA---GACACGCCUCGAACUGAACCGAAAUGUAUUCAUUUCGGUACUCG------AAUUCACCAAUUCAGAUGGUGCCAGUUUUUGUUAUCCGGAGUC----------------- .((....---((((......((((((.(((((((((....))))))))).....------....(((((.......))))).)))))).))))....))....----------------- ( -22.90) >DroYak_CAF1 130302 100 + 1 CCACCCA---GACACGCCUCAAACUGAAGCGAAAUGUAUUCAUUUCGAUACUCGAACUCGAAUACACCAAUUCAGAUGGUGCCAGUUUUUGUUAUCCGGGGUC----------------- ...(((.---((...((...((((((...(((((((....)))))))....(((....)))...(((((.......))))).))))))..))..)).)))...----------------- ( -24.50) >DroAna_CAF1 112541 106 + 1 CCCCCCACUCGUCAUGCCACAAACUGGGCCAAAAUGUAUUCAUUUCAACACU--------------CCAAUGCGAACGGAGGGAGUUUUUGUUAUACCGAGUCCGGCUCUGGGGUCUGGG ...((((((((..(((.((.(((((...((.(((((....))))).....((--------------((.........))))))))))).)).)))..)))))..((((....)))).))) ( -25.50) >consensus CCCCCCA___GACACGCCUCGAACUGAACCGAAAUGUAUUCAUUUCGGUACUCG______AAUACACCAAUUCAGAUGGUGCCAGUUUUUGUUAUCCGGGGUC_________________ ...............((((((......(((((((((....)))))))))...............(((((.......)))))...............)))))).................. (-17.13 = -18.85 + 1.72)

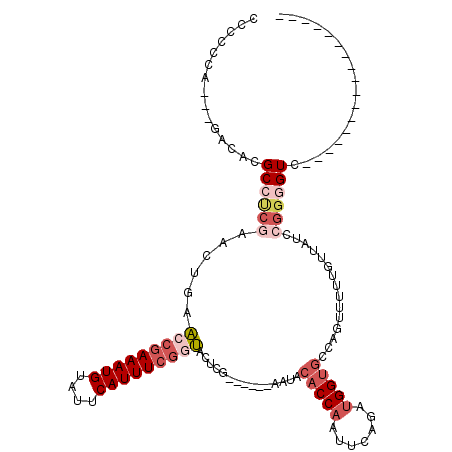

| Location | 21,643,709 – 21,643,803 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.35 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -22.12 |
| Energy contribution | -22.68 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21643709 94 - 27905053 -----------------GACCCCGGAUAACAAAAACUGGCACCAUCUGAAUUGGUGUAUU------CGAGUACCGAAAUGAAUACAUUUCGGUUCACUUCGAGGCGUGUC---CGGGGGG -----------------..(((((((((..........((((((.......)))))).((------((((((((((((((....)))))))))..)).)))))...))))---))))).. ( -42.00) >DroSec_CAF1 133021 94 - 1 -----------------GACCCCGGAUAACAAAAACUGGCACCAUCUGAAUUGGUGUAUU------CGAGUACCGAAAUGAAUACAUUUCGGUUCACUUCGGGGCGUGUC---UGGGGGG -----------------..(((((((((..........((((((.......)))))).((------((((((((((((((....)))))))))..)).)))))...))))---))))).. ( -38.80) >DroSim_CAF1 129633 94 - 1 -----------------GACCCCGGAUAACAAAAACUGGCACCAUCUGAAUUGGUGUAUU------CGAGUACCGAAAUGAAUACAUUUCGGUUCACUUCGGGGCGUGUC---UGGGGGG -----------------..(((((((((..........((((((.......)))))).((------((((((((((((((....)))))))))..)).)))))...))))---))))).. ( -38.80) >DroEre_CAF1 129775 94 - 1 -----------------GACUCCGGAUAACAAAAACUGGCACCAUCUGAAUUGGUGAAUU------CGAGUACCGAAAUGAAUACAUUUCGGUUCAGUUCGAGGCGUGUC---UGUGGGG -----------------..(((((((((...........(((((.......)))))..((------((((.(((((((((....)))))))))....))))))...))))---)).))). ( -31.10) >DroYak_CAF1 130302 100 - 1 -----------------GACCCCGGAUAACAAAAACUGGCACCAUCUGAAUUGGUGUAUUCGAGUUCGAGUAUCGAAAUGAAUACAUUUCGCUUCAGUUUGAGGCGUGUC---UGGGUGG -----------------..(((((((((.(..((((((((((((.......))))))(((((....)))))..(((((((....)))))))...))))))..)...))))---)))).). ( -36.10) >DroAna_CAF1 112541 106 - 1 CCCAGACCCCAGAGCCGGACUCGGUAUAACAAAAACUCCCUCCGUUCGCAUUGG--------------AGUGUUGAAAUGAAUACAUUUUGGCCCAGUUUGUGGCAUGACGAGUGGGGGG (((...((((......))(((((.(((..((.(((((..(((((.......)))--------------)).((..(((((....)))))..))..))))).))..))).))))))).))) ( -30.80) >consensus _________________GACCCCGGAUAACAAAAACUGGCACCAUCUGAAUUGGUGUAUU______CGAGUACCGAAAUGAAUACAUUUCGGUUCACUUCGAGGCGUGUC___UGGGGGG ...................(((((....(((....((.((((((.......)))))).........((((.(((((((((....)))))))))....))))))...)))....))))).. (-22.12 = -22.68 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:41 2006