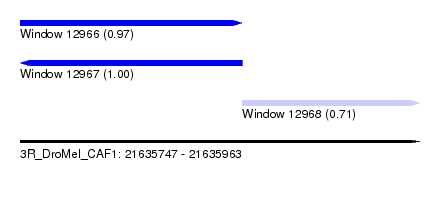
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,635,747 – 21,635,963 |
| Length | 216 |
| Max. P | 0.997400 |

| Location | 21,635,747 – 21,635,867 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -27.04 |
| Energy contribution | -27.12 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21635747 120 + 27905053 CAUUUCGAUUGCGGCUCGGCUGUUGCUAUAUCGUCCUGUUUAUGCCUGAUUAAAGUCAAACAACAAACAUAUUCUGAGCUGCAUUUACCAGACAAUCGUAUAAACAUAUACACAUGUGGU ......((.(((((((((((....))).........(((((.((..((((....))))..))..)))))......)))))))).))((((.(.....(((((....)))))...).)))) ( -25.20) >DroSec_CAF1 125076 120 + 1 CAUUUCGAUUGCGGCUCGGCUGUUGCCCUAUCGUCCUGUUUAUGCCUGAUUAAAGUCAAACAACAAACAUAUUCUGAGCUGCAUUUACCAGACAAUCGUAUAAACAUAUACACUUGUGGU ......((.(((((((((((....))).........(((((.((..((((....))))..))..)))))......)))))))).))(((..((((..(((((....)))))..))))))) ( -28.30) >DroSim_CAF1 121684 120 + 1 CAUUUCGAUUGCGGCUCGGCUGUUGCCCUAUCGUCCUGUUUAUGCCUGAUUAAAGUCAAACAACAAACAUAUUCUGAGCUGCAUUUACCAGACAAUCGUAUAAACAUAUACACUUGUGGU ......((.(((((((((((....))).........(((((.((..((((....))))..))..)))))......)))))))).))(((..((((..(((((....)))))..))))))) ( -28.30) >DroEre_CAF1 117541 117 + 1 CAUUUCGAUUGCGGCUCGGCUGUUGCCCUAU---CCUGUUUAUGCCUGAUUAAAGUCAAACAACAAACAUAUUCUGAGCUGCAUUUACCAGACAAUCGUAUAAACAUAUACACUUGUGGU ......((.(((((((((((....)))....---..(((((.((..((((....))))..))..)))))......)))))))).))(((..((((..(((((....)))))..))))))) ( -28.30) >DroYak_CAF1 122203 120 + 1 CAUUUCGAUUGCGGCUCGGCUGUUGCCCUAUCGUCCUGUUUAUGCCUGAUUAAAGUCAAACAACAAACAUAUACUGAGCUGCAUUUGCCAGACAAUCGUAUAAACAUAUACACAUGUGGU .....(((.(((((((((((....))).........(((((.((..((((....))))..))..)))))......)))))))).)))(((.(.....(((((....)))))...).))). ( -27.30) >consensus CAUUUCGAUUGCGGCUCGGCUGUUGCCCUAUCGUCCUGUUUAUGCCUGAUUAAAGUCAAACAACAAACAUAUUCUGAGCUGCAUUUACCAGACAAUCGUAUAAACAUAUACACUUGUGGU ......((.(((((((((((....))).........(((((.((..((((....))))..))..)))))......)))))))).))(((..((((..(((((....)))))..))))))) (-27.04 = -27.12 + 0.08)

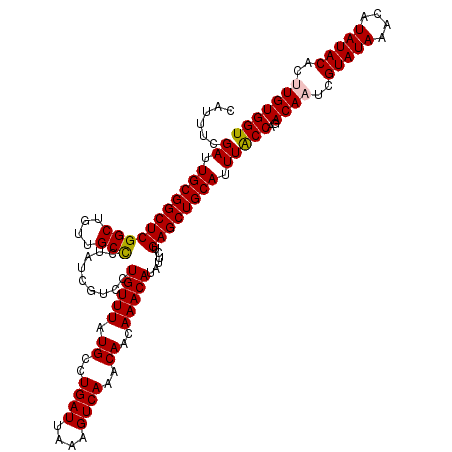
| Location | 21,635,747 – 21,635,867 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -31.36 |
| Energy contribution | -32.16 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21635747 120 - 27905053 ACCACAUGUGUAUAUGUUUAUACGAUUGUCUGGUAAAUGCAGCUCAGAAUAUGUUUGUUGUUUGACUUUAAUCAGGCAUAAACAGGACGAUAUAGCAACAGCCGAGCCGCAAUCGAAAUG ((((.((.((((((....))))))...)).))))...(((.((((......(((((((.((((((......)))))))))))))..........((....)).)))).)))......... ( -28.80) >DroSec_CAF1 125076 120 - 1 ACCACAAGUGUAUAUGUUUAUACGAUUGUCUGGUAAAUGCAGCUCAGAAUAUGUUUGUUGUUUGACUUUAAUCAGGCAUAAACAGGACGAUAGGGCAACAGCCGAGCCGCAAUCGAAAUG (((((((.((((((....)))))).))))..)))...(((.((((......(((((((.((((((......))))))))))))).........(((....))))))).)))......... ( -34.20) >DroSim_CAF1 121684 120 - 1 ACCACAAGUGUAUAUGUUUAUACGAUUGUCUGGUAAAUGCAGCUCAGAAUAUGUUUGUUGUUUGACUUUAAUCAGGCAUAAACAGGACGAUAGGGCAACAGCCGAGCCGCAAUCGAAAUG (((((((.((((((....)))))).))))..)))...(((.((((......(((((((.((((((......))))))))))))).........(((....))))))).)))......... ( -34.20) >DroEre_CAF1 117541 117 - 1 ACCACAAGUGUAUAUGUUUAUACGAUUGUCUGGUAAAUGCAGCUCAGAAUAUGUUUGUUGUUUGACUUUAAUCAGGCAUAAACAGG---AUAGGGCAACAGCCGAGCCGCAAUCGAAAUG (((((((.((((((....)))))).))))..)))...(((.((((......(((((((.((((((......)))))))))))))..---....(((....))))))).)))......... ( -34.20) >DroYak_CAF1 122203 120 - 1 ACCACAUGUGUAUAUGUUUAUACGAUUGUCUGGCAAAUGCAGCUCAGUAUAUGUUUGUUGUUUGACUUUAAUCAGGCAUAAACAGGACGAUAGGGCAACAGCCGAGCCGCAAUCGAAAUG .......(((((......)))))((((((...((....)).((((.((...(((((((.((((((......)))))))))))))..)).....(((....))))))).))))))...... ( -33.70) >consensus ACCACAAGUGUAUAUGUUUAUACGAUUGUCUGGUAAAUGCAGCUCAGAAUAUGUUUGUUGUUUGACUUUAAUCAGGCAUAAACAGGACGAUAGGGCAACAGCCGAGCCGCAAUCGAAAUG (((((((.((((((....)))))).))))..)))...(((.((((......(((((((.((((((......))))))))))))).........(((....))))))).)))......... (-31.36 = -32.16 + 0.80)



| Location | 21,635,867 – 21,635,963 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 73.46 |
| Mean single sequence MFE | -16.67 |
| Consensus MFE | -2.28 |
| Energy contribution | -5.08 |
| Covariance contribution | 2.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.14 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21635867 96 + 27905053 CAUCAGAAUAGAAUU-----UAUUAGUUGGGAGAUCU---------ACAAAUCUGCCAAUUAUUUUAAGCAAAUUGUUUUUACUCUCCUU-UAACGCAAACAACUUUUAUU ......(((((((((-----((.(((((((.((((..---------....)))).)))))))...))))....((((((...........-......)))))).))))))) ( -13.33) >DroSec_CAF1 125196 100 + 1 CAUAAGAAUAGAAUUCCU--UAUUAGUUGGCAGAUCU---------ACACAUCUGCCAAGUAUUUCAAGCAAAUUGUUUUUACUCCCCUUUUUAAACAAACAACUUUUGUU ..(((((((((.....((--(..((.(((((((((..---------....))))))))).))....)))....)))))))))............((((((.....)))))) ( -19.20) >DroSim_CAF1 121804 100 + 1 CAUAAGAAUAGAAUUCCU--UAUUAGUUGGCAGAUCU---------ACAUAUAUGCCAAUUAUUUCAAGCAAAUUGUUUUUACUCCCCUUUUUAAACAAACAACUUUUGUU ..(((((((((.....((--(..(((((((((.....---------.......)))))))))....)))....)))))))))............((((((.....)))))) ( -14.90) >DroEre_CAF1 117658 109 + 1 CAUAAGAAUAGAAUUGCUCGAAAUAGUCUCGAGAAUUAAUUCAAACACAAGUCUGCCAAUUAUUUCUGGCAAAUUGUU-UUACUUAG-UUCUGAAAUACACAACUUUUGUU ....(((((.(((((((((((.......)))))...))))))....((((...(((((........)))))..)))).-.......)-))))................... ( -19.90) >DroYak_CAF1 122323 104 + 1 CAUAAGAAUAGAAUUGUC--UAUUAGGCCCGAGAAUUAAUUCAAAGACAAAUCUGACAAUUAGUUCAAGGAAAUUGUUAUUACUAAG-UUUGAAAAUAAACU----AUGUA ......((((((....))--))))....((..(((((((((...(((....)))...)))))))))..))..........(((..((-((((....))))))----..))) ( -16.00) >consensus CAUAAGAAUAGAAUUCCU__UAUUAGUUGGGAGAUCU_________ACAAAUCUGCCAAUUAUUUCAAGCAAAUUGUUUUUACUCACCUUUUAAAACAAACAACUUUUGUU ..(((((((((............(((((((.((((...............)))).)))))))...........)))))))))............................. ( -2.28 = -5.08 + 2.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:29 2006