
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,632,963 – 21,633,119 |
| Length | 156 |
| Max. P | 0.994311 |

| Location | 21,632,963 – 21,633,079 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -28.66 |
| Energy contribution | -29.02 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21632963 116 + 27905053 GCAUGCAUUUCACAUGAGCCGGCAGUUGCCGCUUUCCAUUUGGUUUAUUGCGAAUUUUAUUGUUUUACGGUAGUUUAUGGCAAAUGCCAUACAACGAGCCGUGUAAAUAUUUG----UUC (((......(((.((((((.(((....))))))...))).))).....)))((((..((((....((((((.(((((((((....)))))).)))..))))))..))))...)----))) ( -33.10) >DroSec_CAF1 122420 116 + 1 GCAUGCAUUUCACAUGAGCCGGCAGUUGCCGCUUUCCAUUUGGUUUAUUGCGAAUUUUAUUGUUUUACGGUAGUUUAUGGCAAAUGCCAUACAACGAGCCGUGUAAAUAUUUG----UUC (((......(((.((((((.(((....))))))...))).))).....)))((((..((((....((((((.(((((((((....)))))).)))..))))))..))))...)----))) ( -33.10) >DroSim_CAF1 118964 116 + 1 GCCUGCAUUUCACAUGAGCCGGCAGUUGCCGCUUUCCAUUUGGUUUAUUGCGAAUUUUAUUGUUUUACGGUAGUUUAUGGCAAAUGCCAUACAACGAGCCGUGUAAAUAUUUG----UUC ....(((..(((.((((((.(((....))))))...))).))).....)))((((..((((....((((((.(((((((((....)))))).)))..))))))..))))...)----))) ( -32.50) >DroEre_CAF1 114650 116 + 1 GCAUGCAUUUCACAUGAGCCGGCAGUUGCCGCUUUCCAUUUGGUUUAUUGCGAAUUUUAUUGUUUUACGGUAGUUUAUGGCAAAUGCCAUACAACGAGCCGUGUAAAUAUUUG----UUC (((......(((.((((((.(((....))))))...))).))).....)))((((..((((....((((((.(((((((((....)))))).)))..))))))..))))...)----))) ( -33.10) >DroYak_CAF1 119341 116 + 1 GCAUGCAUUUCACAUGAGCCGGCAGUUGCCGCUUUCCAUUUGGUUUAUUGCGAAUUUUAUUGUUUUACGGUAGUUUAUGGCAAAUGCCAUACAACGAGCCGUGUAAAUAUUUG----UUC (((......(((.((((((.(((....))))))...))).))).....)))((((..((((....((((((.(((((((((....)))))).)))..))))))..))))...)----))) ( -33.10) >DroAna_CAF1 101533 120 + 1 GCAUGCAUUUCACAUGAGACGGCAGUUUCCGCUUUCCAUUUGGUUUAUUGCGAAUUUUAUUGUUUAAUGGUAGUUUAUGGCAAUCGCCAUACACCAAGCCGUGUAAAUAUUUGUCUGUUC .((((.......))))((((((((((..(((.........)))...))))).........(((((((((((.((.((((((....)))))).))...))))).))))))..))))).... ( -28.20) >consensus GCAUGCAUUUCACAUGAGCCGGCAGUUGCCGCUUUCCAUUUGGUUUAUUGCGAAUUUUAUUGUUUUACGGUAGUUUAUGGCAAAUGCCAUACAACGAGCCGUGUAAAUAUUUG____UUC ...((((..(((.(((((.((((....))))))...))).))).....))))........(((((((((((.(((((((((....)))))).)))..)))))).)))))........... (-28.66 = -29.02 + 0.36)

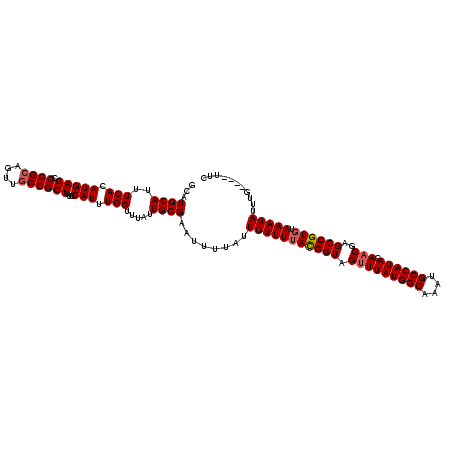

| Location | 21,632,963 – 21,633,079 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -24.85 |
| Energy contribution | -25.35 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21632963 116 - 27905053 GAA----CAAAUAUUUACACGGCUCGUUGUAUGGCAUUUGCCAUAAACUACCGUAAAACAAUAAAAUUCGCAAUAAACCAAAUGGAAAGCGGCAACUGCCGGCUCAUGUGAAAUGCAUGC ...----...........((((...(((.((((((....)))))))))..))))............((((((.....((....))..((((((....))).)))..))))))........ ( -27.60) >DroSec_CAF1 122420 116 - 1 GAA----CAAAUAUUUACACGGCUCGUUGUAUGGCAUUUGCCAUAAACUACCGUAAAACAAUAAAAUUCGCAAUAAACCAAAUGGAAAGCGGCAACUGCCGGCUCAUGUGAAAUGCAUGC ...----...........((((...(((.((((((....)))))))))..))))............((((((.....((....))..((((((....))).)))..))))))........ ( -27.60) >DroSim_CAF1 118964 116 - 1 GAA----CAAAUAUUUACACGGCUCGUUGUAUGGCAUUUGCCAUAAACUACCGUAAAACAAUAAAAUUCGCAAUAAACCAAAUGGAAAGCGGCAACUGCCGGCUCAUGUGAAAUGCAGGC ...----...........((((...(((.((((((....)))))))))..))))............((((((.....((....))..((((((....))).)))..))))))........ ( -27.60) >DroEre_CAF1 114650 116 - 1 GAA----CAAAUAUUUACACGGCUCGUUGUAUGGCAUUUGCCAUAAACUACCGUAAAACAAUAAAAUUCGCAAUAAACCAAAUGGAAAGCGGCAACUGCCGGCUCAUGUGAAAUGCAUGC ...----...........((((...(((.((((((....)))))))))..))))............((((((.....((....))..((((((....))).)))..))))))........ ( -27.60) >DroYak_CAF1 119341 116 - 1 GAA----CAAAUAUUUACACGGCUCGUUGUAUGGCAUUUGCCAUAAACUACCGUAAAACAAUAAAAUUCGCAAUAAACCAAAUGGAAAGCGGCAACUGCCGGCUCAUGUGAAAUGCAUGC ...----...........((((...(((.((((((....)))))))))..))))............((((((.....((....))..((((((....))).)))..))))))........ ( -27.60) >DroAna_CAF1 101533 120 - 1 GAACAGACAAAUAUUUACACGGCUUGGUGUAUGGCGAUUGCCAUAAACUACCAUUAAACAAUAAAAUUCGCAAUAAACCAAAUGGAAAGCGGAAACUGCCGUCUCAUGUGAAAUGCAUGC ..................(((((.(((((((((((....))))))...))))).............(((((......((....))...)))))....)))))..(((((.....))))). ( -27.40) >consensus GAA____CAAAUAUUUACACGGCUCGUUGUAUGGCAUUUGCCAUAAACUACCGUAAAACAAUAAAAUUCGCAAUAAACCAAAUGGAAAGCGGCAACUGCCGGCUCAUGUGAAAUGCAUGC ..................((((...(((.((((((....)))))))))..))))............((((((.....((....))..((((((....)))).))..))))))........ (-24.85 = -25.35 + 0.50)


| Location | 21,633,003 – 21,633,119 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 98.62 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -29.48 |
| Energy contribution | -29.68 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21633003 116 + 27905053 UGGUUUAUUGCGAAUUUUAUUGUUUUACGGUAGUUUAUGGCAAAUGCCAUACAACGAGCCGUGUAAAUAUUUGUUCUAGGCCGUGUCCCCCACAAUUCCCUUUGCAUUUUGCCCAG .(((..(.(((((((..((((....((((((.(((((((((....)))))).)))..))))))..))))...))))......(((.....)))..........))).)..)))... ( -30.40) >DroSec_CAF1 122460 116 + 1 UGGUUUAUUGCGAAUUUUAUUGUUUUACGGUAGUUUAUGGCAAAUGCCAUACAACGAGCCGUGUAAAUAUUUGUUCUAGGCCGUGUCCCCCACAAUUCCCUUUGCAUUUUGCCCAG .(((..(.(((((((..((((....((((((.(((((((((....)))))).)))..))))))..))))...))))......(((.....)))..........))).)..)))... ( -30.40) >DroSim_CAF1 119004 116 + 1 UGGUUUAUUGCGAAUUUUAUUGUUUUACGGUAGUUUAUGGCAAAUGCCAUACAACGAGCCGUGUAAAUAUUUGUUCUAGGCCGUGUCCCCCACAAUUCCCUUUGCAUUUUGCCCAG .(((..(.(((((((..((((....((((((.(((((((((....)))))).)))..))))))..))))...))))......(((.....)))..........))).)..)))... ( -30.40) >DroEre_CAF1 114690 116 + 1 UGGUUUAUUGCGAAUUUUAUUGUUUUACGGUAGUUUAUGGCAAAUGCCAUACAACGAGCCGUGUAAAUAUUUGUUCUAGGCCGUGUCCCCCACAAUUCCCUUUGCAGUUUGCCCAG .(((..(((((((((..((((....((((((.(((((((((....)))))).)))..))))))..))))...))))......(((.....)))..........)))))..)))... ( -33.60) >DroYak_CAF1 119381 116 + 1 UGGUUUAUUGCGAAUUUUAUUGUUUUACGGUAGUUUAUGGCAAAUGCCAUACAACGAGCCGUGUAAAUAUUUGUUCUAGGCCGUGUCCCCCUCAAUUCCCUUUGCAUUUCGCCCGG .((((((..(((((...((((....((((((.(((((((((....)))))).)))..))))))..)))))))))..))))))(((.......(((......))).....))).... ( -28.80) >consensus UGGUUUAUUGCGAAUUUUAUUGUUUUACGGUAGUUUAUGGCAAAUGCCAUACAACGAGCCGUGUAAAUAUUUGUUCUAGGCCGUGUCCCCCACAAUUCCCUUUGCAUUUUGCCCAG .(((..(.(((((((..((((....((((((.(((((((((....)))))).)))..))))))..))))...)))).(((...(((.....)))....)))..))).)..)))... (-29.48 = -29.68 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:25 2006