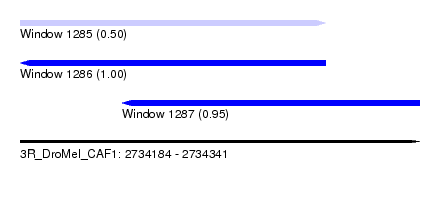
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,734,184 – 2,734,341 |
| Length | 157 |
| Max. P | 0.999130 |

| Location | 2,734,184 – 2,734,304 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -27.50 |
| Energy contribution | -27.62 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.91 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2734184 120 + 27905053 AAGAAGAAAAAUUCCACUUAAUACGUUUCGUGGAAUUGUCGAUCACUUUCCUUGACACUUACCAAACUGACUUCGCAUCCAGGGAUACAGUGGAGACGGCAUCCCCGAGGACUGCGAGUU ...(((...((((((((............))))))))(((((.........))))).)))........((((.(((((((.(((((.(...(....).).)))))...))).)))))))) ( -30.90) >DroPse_CAF1 29815 120 + 1 AAGAAGAAAAAUUCCACUUAAUACGUUUCGUGGAAUUAUCGAUCACUUUCCUUGACACUUACCAAACUGACUUCGCAUCCAGGGAUACAGUGGUGACGGCAUCCCCGAGGAUUGCGAAUU ..((.((..((((((((............)))))))).))..))...........................(((((((((.(((((.(...(....).).)))))...))).)))))).. ( -27.50) >DroGri_CAF1 48057 120 + 1 AAGAAGAAAAAUUCCACUUAAUACGUUUCGUGGAAUUGUCGAUCACUUUCCUUGACACUUACCAAACUGACUUCGCAUCCAGGGAUACAGUGGUGACGGCAUCCCUGAGGAUUGCGAACU ...(((...((((((((............))))))))(((((.........))))).)))...........(((((((((((((((.(...(....).).))))))..))).)))))).. ( -31.90) >DroWil_CAF1 21255 120 + 1 AAGAAGAAAAAUUCCACUUAAUACGUUUCGUGGAAUUGUCGAUCACUUUCCUUGACACUUACCAAACUGACUUCGCAUCCAGGGAUACAGUGGGGACGGCAUCCCUGAGGAUUGCGAAUU ...(((...((((((((............))))))))(((((.........))))).)))...........(((((((((((((((.(...(....).).))))))..))).)))))).. ( -32.00) >DroAna_CAF1 9820 119 + 1 AAGAAGAAAAAUUCCACUUAAUACGU-UCGUGGAAUUGUCGAUCACUUUCCUUGACACUUACCAAACUGACUUCGCAUCCAGGGAUACAGUGGUGACGGCAUCCCCGAGGAUUGCGAGUU ...(((...((((((((.........-..))))))))(((((.........))))).)))........((((.(((((((.(((((.(...(....).).)))))...))).)))))))) ( -31.00) >DroPer_CAF1 28877 120 + 1 AAGAAGAAAAAUUCCACUUAAUACGUUUCGUGGAAUUAUCGAUCACUUUCCUUGACACUUACCAAACUGACUUCGCAUCCAGGGAUACAGUGGUGACGGCAUCCCCGAGGAUUGCGAAUU ..((.((..((((((((............)))))))).))..))...........................(((((((((.(((((.(...(....).).)))))...))).)))))).. ( -27.50) >consensus AAGAAGAAAAAUUCCACUUAAUACGUUUCGUGGAAUUGUCGAUCACUUUCCUUGACACUUACCAAACUGACUUCGCAUCCAGGGAUACAGUGGUGACGGCAUCCCCGAGGAUUGCGAAUU ...(((...((((((((............))))))))(((((.........))))).)))...........(((((((((.(((((.(.((....)).).)))))...))).)))))).. (-27.50 = -27.62 + 0.11)

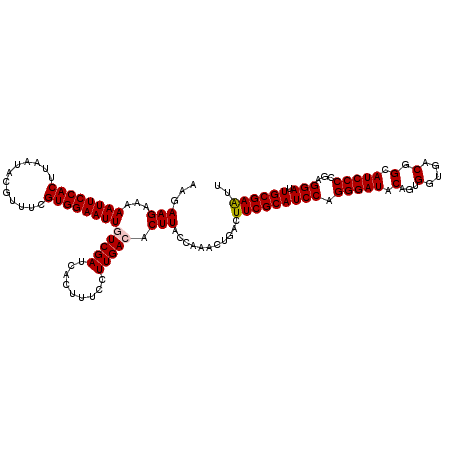
| Location | 2,734,184 – 2,734,304 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -35.13 |
| Consensus MFE | -35.15 |
| Energy contribution | -34.93 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.66 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.999130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2734184 120 - 27905053 AACUCGCAGUCCUCGGGGAUGCCGUCUCCACUGUAUCCCUGGAUGCGAAGUCAGUUUGGUAAGUGUCAAGGAAAGUGAUCGACAAUUCCACGAAACGUAUUAAGUGGAAUUUUUCUUCUU ...((((..(((((((((((((.((....)).)))))))))(((((....((.....))...))))).))))..))))..((.((((((((............))))))))..))..... ( -35.30) >DroPse_CAF1 29815 120 - 1 AAUUCGCAAUCCUCGGGGAUGCCGUCACCACUGUAUCCCUGGAUGCGAAGUCAGUUUGGUAAGUGUCAAGGAAAGUGAUCGAUAAUUCCACGAAACGUAUUAAGUGGAAUUUUUCUUCUU ...((((..(((((((((((((.((....)).)))))))))(((((....((.....))...))))).))))..))))..((.((((((((............))))))))..))..... ( -35.20) >DroGri_CAF1 48057 120 - 1 AGUUCGCAAUCCUCAGGGAUGCCGUCACCACUGUAUCCCUGGAUGCGAAGUCAGUUUGGUAAGUGUCAAGGAAAGUGAUCGACAAUUCCACGAAACGUAUUAAGUGGAAUUUUUCUUCUU .(.((((..(((((((((((((.((....)).)))))))))(((((....((.....))...))))).))))..)))).)((.((((((((............))))))))..))..... ( -35.50) >DroWil_CAF1 21255 120 - 1 AAUUCGCAAUCCUCAGGGAUGCCGUCCCCACUGUAUCCCUGGAUGCGAAGUCAGUUUGGUAAGUGUCAAGGAAAGUGAUCGACAAUUCCACGAAACGUAUUAAGUGGAAUUUUUCUUCUU ..((((((.(((..((((((((.((....)).)))))))))))))))))(((..(((((......)))))((......)))))((((((((............))))))))......... ( -35.30) >DroAna_CAF1 9820 119 - 1 AACUCGCAAUCCUCGGGGAUGCCGUCACCACUGUAUCCCUGGAUGCGAAGUCAGUUUGGUAAGUGUCAAGGAAAGUGAUCGACAAUUCCACGA-ACGUAUUAAGUGGAAUUUUUCUUCUU ...(((((.(((..((((((((.((....)).)))))))))))))))).(((..(((((......)))))((......)))))((((((((..-.........))))))))......... ( -34.30) >DroPer_CAF1 28877 120 - 1 AAUUCGCAAUCCUCGGGGAUGCCGUCACCACUGUAUCCCUGGAUGCGAAGUCAGUUUGGUAAGUGUCAAGGAAAGUGAUCGAUAAUUCCACGAAACGUAUUAAGUGGAAUUUUUCUUCUU ...((((..(((((((((((((.((....)).)))))))))(((((....((.....))...))))).))))..))))..((.((((((((............))))))))..))..... ( -35.20) >consensus AAUUCGCAAUCCUCGGGGAUGCCGUCACCACUGUAUCCCUGGAUGCGAAGUCAGUUUGGUAAGUGUCAAGGAAAGUGAUCGACAAUUCCACGAAACGUAUUAAGUGGAAUUUUUCUUCUU ...((((..(((((((((((((.((....)).)))))))))(((((....((.....))...))))).))))..))))..((.((((((((............))))))))..))..... (-35.15 = -34.93 + -0.22)

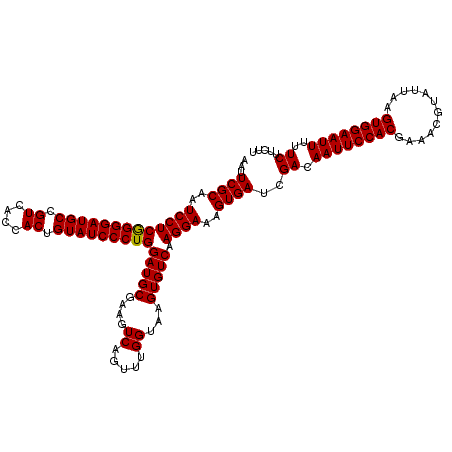
| Location | 2,734,224 – 2,734,341 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -37.44 |
| Consensus MFE | -32.74 |
| Energy contribution | -32.38 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.950980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2734224 117 - 27905053 ACAUC---CUGGCCAACACACGCCUCCUUCCCAAAACCCGAACUCGCAGUCCUCGGGGAUGCCGUCUCCACUGUAUCCCUGGAUGCGAAGUCAGUUUGGUAAGUGUCAAGGAAAGUGAUC ..(((---(((((........)))(((((..((....(((((((((((.(((..((((((((.((....)).)))))))))))))))).....))))))....))..))))).)).))). ( -39.10) >DroVir_CAF1 49555 117 - 1 ACAUC---CCGGACAACAUACACCGCCAUCCCAAAAUCCGAGUUCGCAAUCCUCAGGGAUGCCGUCUCCACUGUAUCCCUGGAUGCGAAGUCAGUUUGGUAAGUGUCAAGGAAAGUGAUC ...((---(..((((...............(((((....((.((((((.(((..((((((((.((....)).))))))))))))))))).))..)))))....))))..)))........ ( -37.31) >DroPse_CAF1 29855 120 - 1 CCAUUUAUCGGGCCAACAUACACCGCCAUCCCAAAACCCGAAUUCGCAAUCCUCGGGGAUGCCGUCACCACUGUAUCCCUGGAUGCGAAGUCAGUUUGGUAAGUGUCAAGGAAAGUGAUC .((((((((.(((...........)))......((((..((.((((((.(((..((((((((.((....)).))))))))))))))))).)).))))))))))))............... ( -36.30) >DroWil_CAF1 21295 117 - 1 ACAUC---CAGGACAACACACACCGCCAUCCCAAAAUCCGAAUUCGCAAUCCUCAGGGAUGCCGUCCCCACUGUAUCCCUGGAUGCGAAGUCAGUUUGGUAAGUGUCAAGGAAAGUGAUC ...((---(..((((...............(((((....((.((((((.(((..((((((((.((....)).))))))))))))))))).))..)))))....))))..)))........ ( -35.11) >DroMoj_CAF1 51410 117 - 1 ACAUC---CCGGACAGCAUACGCCUCCAUCCCAAAAUCCGAGUUCGCAGUCCUCAGGGAUGCCGUCUCCACUGUAUCCCUGGAUGCGAAGUCAGUUUGGUAAGUGUCAAGGAAAGUGAUC ...((---(..((((((....)).......(((((....((.((((((....((((((((((.((....)).)))))))))).)))))).))..)))))....))))..)))........ ( -40.50) >DroPer_CAF1 28917 120 - 1 CCAUUUAUCGGGCCAACAUACACCGCCAUCCCAAAACCCGAAUUCGCAAUCCUCGGGGAUGCCGUCACCACUGUAUCCCUGGAUGCGAAGUCAGUUUGGUAAGUGUCAAGGAAAGUGAUC .((((((((.(((...........)))......((((..((.((((((.(((..((((((((.((....)).))))))))))))))))).)).))))))))))))............... ( -36.30) >consensus ACAUC___CCGGACAACAUACACCGCCAUCCCAAAACCCGAAUUCGCAAUCCUCAGGGAUGCCGUCUCCACUGUAUCCCUGGAUGCGAAGUCAGUUUGGUAAGUGUCAAGGAAAGUGAUC ................(((...((..(((.(((((....((.(((((.((((..((((((((.((....)).))))))))))))))))).))..)))))...)))....))...)))... (-32.74 = -32.38 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:39 2006