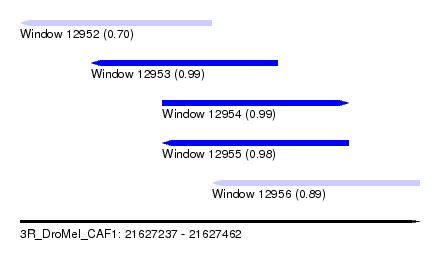
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,627,237 – 21,627,462 |
| Length | 225 |
| Max. P | 0.993066 |

| Location | 21,627,237 – 21,627,345 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.75 |
| Mean single sequence MFE | -22.20 |
| Consensus MFE | -15.50 |
| Energy contribution | -15.82 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21627237 108 - 27905053 ----------AAAA-CGACGGCAAACCAAGUGUCAAGUUGAAAUAAACUUUAAGACACACUUGACAUAUUGCCAGUUGCUCGCUUCCAGCUCACAACAUAAUUUUCCAUUAGUUGGAUA ----------....-(((((((((.....(((((((((((...............)).))))))))).))))).((((...((.....))...))))..............)))).... ( -23.66) >DroSec_CAF1 116643 108 - 1 ----------AAAA-CGACGGCAAACCAAGUGUCAAGUUGAAAUAAACUUUAAGACAGACUUGACAUAUUGCCAGUUGCUCGCUUCCAGCUCACAACAUAAUUUUCCAUUAGUUGGAUA ----------....-(((((((((.....((((((((((..................)))))))))).))))).((((...((.....))...))))..............)))).... ( -25.27) >DroSim_CAF1 113197 96 - 1 ----------A-------------AUCAAGUGUCAAGUUGAAAUAAACUUUAAGACAGACUUGACAUAUUGCCAGUUGCUCGCUUUCAGCUCACAACAUAAUUUUCCAUUAGUUGGAUA ----------.-------------.....((((((((((..................)))))))))).......((((...((.....))...)))).......((((.....)))).. ( -19.07) >DroYak_CAF1 113415 119 - 1 AAAAAAAAAAAACACCGAUAGCAAACCAAGUGUCAAGUUGAAAUAAACUUUAAGACAGACUUGACAUAUUGCCAGUUGCUCGCUUCCAGUUCACAACAUAAUUUUCCAUUAGUUGGAUA ..............(((((.((((.....((((((((((..................)))))))))).))))..((((...((.....))...))))..............)))))... ( -20.27) >DroAna_CAF1 95743 99 - 1 ----------A---------GCA-GUCCAGCGACAACUUGAAGCAAACUUUAGGACAGACUUGACAUAUUGCUAGUUGCUCGCUUCCAGCUCACAACAUAAUUUUCCAUUAGUUGGAUA ----------.---------...-....((((((((((...(((((...(((((.....)))))....)))))))))).)))))(((((((...................))))))).. ( -22.71) >consensus __________AA_A_CGA__GCAAACCAAGUGUCAAGUUGAAAUAAACUUUAAGACAGACUUGACAUAUUGCCAGUUGCUCGCUUCCAGCUCACAACAUAAUUUUCCAUUAGUUGGAUA .............................((((((((((....(........)....)))))))))).......((((...((.....))...)))).......((((.....)))).. (-15.50 = -15.82 + 0.32)

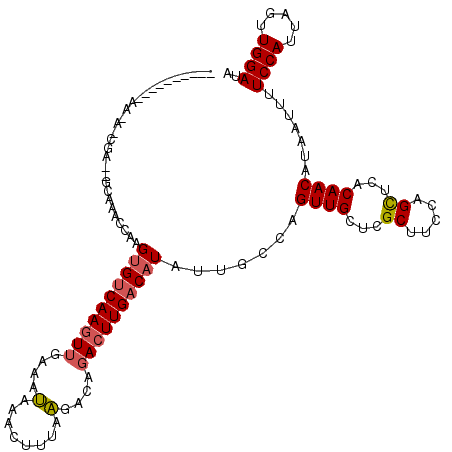

| Location | 21,627,277 – 21,627,382 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.68 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -10.86 |
| Energy contribution | -11.30 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21627277 105 - 27905053 CAAAAAUGCUAUCUCGAAAAAAGAAGGCACAAAAAAA-------------AAAA-CGACGGCAAACCAAGUGUCAAGUUGAAAUAAACUUUAAGACACACUUGACAUAUUGCCAGUUGC ......((((.(((.......))).))))........-------------....-(((((((((.....(((((((((((...............)).))))))))).))))).)))). ( -22.46) >DroSec_CAF1 116683 106 - 1 CAAAAAUGCUAUCUCGCAAAAAGAAGGCAAAAAGAAAA------------AAAA-CGACGGCAAACCAAGUGUCAAGUUGAAAUAAACUUUAAGACAGACUUGACAUAUUGCCAGUUGC ......((((.(((.......))).)))).........------------....-(((((((((.....((((((((((..................)))))))))).))))).)))). ( -24.37) >DroSim_CAF1 113237 94 - 1 CAAAAAUGCUAUCUCGCAAAAAGGAGGCAAAAAGAAAA------------A-------------AUCAAGUGUCAAGUUGAAAUAAACUUUAAGACAGACUUGACAUAUUGCCAGUUGC ......(((......))).......(((((...((...------------.-------------.))..((((((((((..................)))))))))).)))))...... ( -19.57) >DroYak_CAF1 113455 118 - 1 CAAAAAUGCUAUCUCGCAAAAAGA-GGCAAAAAUACAAAAAAAAAAAAAAAACACCGAUAGCAAACCAAGUGUCAAGUUGAAAUAAACUUUAAGACAGACUUGACAUAUUGCCAGUUGC ......(((((((((.......))-((...........................)))))))))......((((((((((..................))))))))))............ ( -18.60) >DroAna_CAF1 95783 96 - 1 CAGAAAUUCUGGCUGGCAGAGAGAAGUGACCAAAAAA-------------A---------GCA-GUCCAGCGACAACUUGAAGCAAACUUUAGGACAGACUUGACAUAUUGCUAGUUGC (((.....)))((((((((.(..((((..........-------------.---------...-(((....)))..(((((((....)))))))....))))..)...))))))))... ( -19.00) >consensus CAAAAAUGCUAUCUCGCAAAAAGAAGGCAAAAAAAAAA____________AA_A_CGA__GCAAACCAAGUGUCAAGUUGAAAUAAACUUUAAGACAGACUUGACAUAUUGCCAGUUGC ......((((.(((.......))).))))........................................((((((((((....(........)....))))))))))............ (-10.86 = -11.30 + 0.44)



| Location | 21,627,317 – 21,627,422 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 68.63 |
| Mean single sequence MFE | -21.87 |
| Consensus MFE | -13.72 |
| Energy contribution | -14.19 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21627317 105 + 27905053 AACUUGACACUUGGUUUGCCGUCGUUUU-UUUUUUUGUGCCUUCUUUUUUCGAGAUAGCAUUUUUGCUGUCGAUUUUUCGAAGGGGAAGAUUUGACAGCUGCACUU .............((..((.((((..((-((((......(((((.........(((((((....)))))))((....)))))))))))))..)))).)).)).... ( -25.60) >DroSim_CAF1 113277 92 + 1 AACUUGACACUUGAU------------UUUUUCUUUUUGCCUCCUUUUUGCGAGAUAGCAUUUUUGCUGUCGAUUUUUCG--GGGGAAGAUUUGACAGCUGCACUU ....((.((.(((.(------------(...((((....(((((.........(((((((....)))))))((....)))--))))))))...))))).))))... ( -20.80) >DroAna_CAF1 95823 81 + 1 AAGUUGUCGCUGGAC-UGC--------U-UUUUUUGGUCACUUCUCUCUGCCAGCCAGAAUUUCUGCUGUCGAUUU---------------UUGACAGCUGCACUU .((((((((.(.(((-.((--------.-..(((((((..(........)...))))))).....)).))).)...---------------.))))))))...... ( -19.20) >consensus AACUUGACACUUGAU_UGC________U_UUUUUUUGUGCCUUCUUUUUGCGAGAUAGCAUUUUUGCUGUCGAUUUUUCG__GGGGAAGAUUUGACAGCUGCACUU .........((((.....................................))))...........((((((((.(((((......))))).))))))))....... (-13.72 = -14.19 + 0.47)

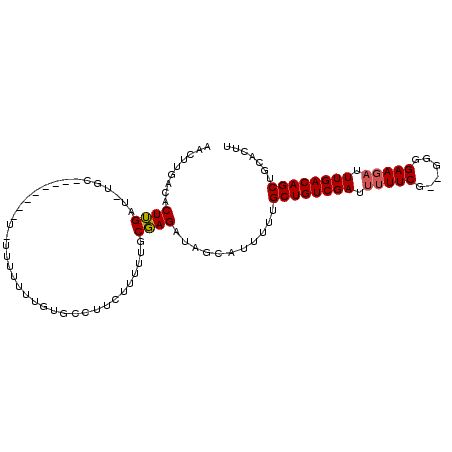

| Location | 21,627,317 – 21,627,422 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 68.63 |
| Mean single sequence MFE | -17.17 |
| Consensus MFE | -11.39 |
| Energy contribution | -11.28 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21627317 105 - 27905053 AAGUGCAGCUGUCAAAUCUUCCCCUUCGAAAAAUCGACAGCAAAAAUGCUAUCUCGAAAAAAGAAGGCACAAAAAAA-AAAACGACGGCAAACCAAGUGUCAAGUU ..((((.((((((.....(((......))).....)))))).......((.(((.......))))))))).......-........((((.......))))..... ( -18.40) >DroSim_CAF1 113277 92 - 1 AAGUGCAGCUGUCAAAUCUUCCCC--CGAAAAAUCGACAGCAAAAAUGCUAUCUCGCAAAAAGGAGGCAAAAAGAAAAA------------AUCAAGUGUCAAGUU ...(((.((((((.....(((...--.))).....)))))).....(((......)))........)))..........------------............... ( -14.70) >DroAna_CAF1 95823 81 - 1 AAGUGCAGCUGUCAA---------------AAAUCGACAGCAGAAAUUCUGGCUGGCAGAGAGAAGUGACCAAAAAA-A--------GCA-GUCCAGCGACAACUU ...(((.((((((..---------------.....)))))).....(((((.....)))))................-.--------)))-(((....)))..... ( -18.40) >consensus AAGUGCAGCUGUCAAAUCUUCCCC__CGAAAAAUCGACAGCAAAAAUGCUAUCUCGCAAAAAGAAGGCACAAAAAAA_A________GCA_AUCAAGUGUCAAGUU ..((((.((((((......................)))))).....((((.(((.......))).))))...........................))).)..... (-11.39 = -11.28 + -0.11)


| Location | 21,627,345 – 21,627,462 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -20.54 |
| Consensus MFE | -13.91 |
| Energy contribution | -14.06 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21627345 117 - 27905053 UCAAGCAAUUGAACAGCGACCAAACUUCCACACAAAUCCCAAGUGCAGCUGUCAAAUCUUCCCCUUCGAAAAAUCGACAGCAAAAAUGCUAUCUCGAAAAAAGAAGGCACAAAAAAA--- ((((....))))(((((......((((.............))))...)))))..........(((((......((((.((((....))))...)))).....)))))..........--- ( -20.62) >DroSec_CAF1 116751 116 - 1 UCAAGCAAUUGAACAGCGACCAAACUUCCACACAAAUCCCAAGUGCAGCUGUCAAAUCUUCCCC--CGAAAAAUCGACAGCAAAAAUGCUAUCUCGCAAAAAGAAGGCAAAAAGAAAA-- ((((....))))...((((...........(((.........)))..((((((.....(((...--.))).....))))))............)))).....................-- ( -18.10) >DroSim_CAF1 113293 116 - 1 UCAAGCAAUUGAACAGCGACCAAACUUCCACACAAAUCCCAAGUGCAGCUGUCAAAUCUUCCCC--CGAAAAAUCGACAGCAAAAAUGCUAUCUCGCAAAAAGGAGGCAAAAAGAAAA-- ((((....))))............(((((..................((((((.....(((...--.))).....)))))).....(((......)))....)))))...........-- ( -20.90) >DroYak_CAF1 113534 117 - 1 UCAAGCAAUUGAACAGCGACCAAACUUCCACACAAAUCCCAAGUGCAGCUGUCAAAUCUUCCUC--CGAAAAAUCGACAGCAAAAAUGCUAUCUCGCAAAAAGA-GGCAAAAAUACAAAA ((((....))))..((((............(((.........)))..((((((.....(((...--.))).....)))))).....)))).((((.......))-))............. ( -18.50) >DroAna_CAF1 95842 100 - 1 UUCAGCAAUUGAACAGGGAA-AAACUU-UGGAGAAGCCCGAAGUGCAGCUGUCAA---------------AAAUCGACAGCAGAAAUUCUGGCUGGCAGAGAGAAGUGACCAAAAAA--- .(((((....(((.......-..((((-(((......)))))))...((((((..---------------.....)))))).....)))..))))).....................--- ( -24.60) >consensus UCAAGCAAUUGAACAGCGACCAAACUUCCACACAAAUCCCAAGUGCAGCUGUCAAAUCUUCCCC__CGAAAAAUCGACAGCAAAAAUGCUAUCUCGCAAAAAGAAGGCAAAAAAAAAA__ ((((....))))...................................((((((.....(((......))).....)))))).....((((.(((.......))).))))........... (-13.91 = -14.06 + 0.15)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:17 2006