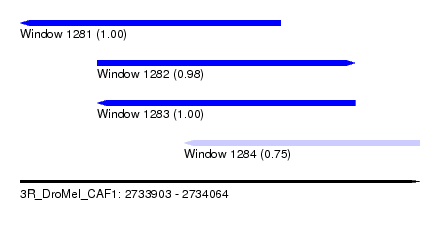
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,733,903 – 2,734,064 |
| Length | 161 |
| Max. P | 0.999586 |

| Location | 2,733,903 – 2,734,008 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.81 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -20.89 |
| Energy contribution | -22.45 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.72 |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2733903 105 - 27905053 ACUGCUUUAUAUUGUGUUUCCUGCUUGAUAUUAGAUCACUAAGCAAGCAG-ACGCGCAAGCAGUUCACGCAGAUCACGCAG---------ACGUUAAAAAUUUAAAAAUGUUUUU .((((......((((((...(((((((...((((....)))).)))))))-..((....)).....)))))).....))))---------......................... ( -27.20) >DroSec_CAF1 9842 106 - 1 ACUGCUUUAUAUUAUGUUUACUGCUUGAUAUUAGAUCACUAAGCAAGCAGUACAUGCAUGCAGCUAGAGUAGAUCGGGCAG---------ACGUUAAAAAUUUAAAAAUGUUUUU .((((((((...(((((.(((((((((...((((....)))).)))))))))...)))))....)))))))).......((---------(((((...........))))))).. ( -28.10) >DroSim_CAF1 10573 106 - 1 ACUGCUUUAUUUUGUACUUACUGCUUGAUAUUAGAUCACUAAGCAAGCAGUGCGUGCAAGCAGUUCGCGAAGAUCGGGCAG---------ACGUUAAGAAUUUAAAAAUGUUUUU .(((((..(((((((((.(((((((((...((((....)))).))))))))).))))..((.....)).)))))..)))))---------......................... ( -32.50) >DroEre_CAF1 11042 105 - 1 ACAGCUUGGUAGCGCGUUUACUGCUUGACAGUAGAUCACAUAGCAAGCAGU-GAUGCGAGCAGUUCAAGUC--------UGCUUGAUAAGACGUUUAACAUUUAAACAU-UUUUG .((((((((..((.(((((((((((((...((.....))....))))))))-)).))).))...))))).)--------))...........(((((.....)))))..-..... ( -28.70) >consensus ACUGCUUUAUAUUGUGUUUACUGCUUGAUAUUAGAUCACUAAGCAAGCAGUACAUGCAAGCAGUUCAAGUAGAUCGGGCAG_________ACGUUAAAAAUUUAAAAAUGUUUUU .((((((((..((((((.(((((((((...((((....)))).))))))))).)))))).....))))))))........................................... (-20.89 = -22.45 + 1.56)


| Location | 2,733,934 – 2,734,038 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.22 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -17.53 |
| Energy contribution | -17.49 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2733934 104 + 27905053 GAUCUGCGUGAACUGCUUGCGCGU-CUGCUUGCUUAGUGAUCUAAUAUCAAGCAGGAAACACAAUAUAAAGCAGUACAUCUAGACUGCUUUUUACUGCUUUGCUU .....(((..(.....)..))).(-(((((((.((((....))))...))))))))..........((((((((((.....((....))...))))))))))... ( -27.90) >DroSec_CAF1 9873 105 + 1 GAUCUACUCUAGCUGCAUGCAUGUACUGCUUGCUUAGUGAUCUAAUAUCAAGCAGUAAACAUAAUAUAAAGCAGUAUAUCUUGACUGCUUCCCACUGCUUUGCUU ..............(((.((((((((((((((.((((....))))...)))))))))...........(((((((........)))))))..)).)))..))).. ( -27.10) >DroSim_CAF1 10604 105 + 1 GAUCUUCGCGAACUGCUUGCACGCACUGCUUGCUUAGUGAUCUAAUAUCAAGCAGUAAGUACAAAAUAAAGCAGUAUAUCUUGACUGCUUCCUACUGCUUUGCUU .......(((((..(((((.((..((((((((.((((....))))...))))))))..)).)))....(((((((........)))))))......))))))).. ( -28.10) >DroEre_CAF1 11077 99 + 1 ----GACUUGAACUGCUCGCAUC-ACUGCUUGCUAUGUGAUCUACUGUCAAGCAGUAAACGCGCUACCAAGCUGUGUAU-UCGACUGCUUUGUACUGCUUUGUUU ----.....((((...(((.(((-((.(((((.((((((...(((((.....)))))..)))).)).))))).))).))-.)))..((........))...)))) ( -22.00) >DroYak_CAF1 9712 104 + 1 GACCUGCUUGACCUGCUUGUAUG-ACUGCUUGCGUAUUGAUCUCGCAUUAAGCAGUAAACACGCUAUCAAGCAGGGGAUUUCGACUGCUUUGUACUGCAUUGCUU ..(((((((((...((.(((...-((((((((((.........))))...))))))..))).))..)))))))))...........((..((.....))..)).. ( -35.80) >consensus GAUCUACUUGAACUGCUUGCAUG_ACUGCUUGCUUAGUGAUCUAAUAUCAAGCAGUAAACACAAUAUAAAGCAGUAUAUCUCGACUGCUUUCUACUGCUUUGCUU ..............((..(((...((((((((................))))))))............(((((((........))))))).....)))...)).. (-17.53 = -17.49 + -0.04)

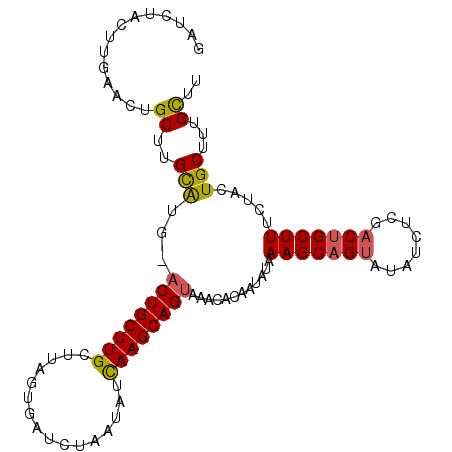
| Location | 2,733,934 – 2,734,038 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.22 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -22.96 |
| Energy contribution | -24.72 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.32 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.71 |
| SVM decision value | 3.75 |
| SVM RNA-class probability | 0.999586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2733934 104 - 27905053 AAGCAAAGCAGUAAAAAGCAGUCUAGAUGUACUGCUUUAUAUUGUGUUUCCUGCUUGAUAUUAGAUCACUAAGCAAGCAG-ACGCGCAAGCAGUUCACGCAGAUC ..((...((((((.(((((((((.....).)))))))).)))))).....(((((((...((((....)))).)))))))-..((....)).......))..... ( -35.40) >DroSec_CAF1 9873 105 - 1 AAGCAAAGCAGUGGGAAGCAGUCAAGAUAUACUGCUUUAUAUUAUGUUUACUGCUUGAUAUUAGAUCACUAAGCAAGCAGUACAUGCAUGCAGCUAGAGUAGAUC .(((...((((((.((((((((........)))))))).))))((((.(((((((((...((((....)))).)))))))))...)))))).))).......... ( -31.60) >DroSim_CAF1 10604 105 - 1 AAGCAAAGCAGUAGGAAGCAGUCAAGAUAUACUGCUUUAUUUUGUACUUACUGCUUGAUAUUAGAUCACUAAGCAAGCAGUGCGUGCAAGCAGUUCGCGAAGAUC ..(((((((((((.....(......)...)))))))))..(((((((.(((((((((...((((....)))).))))))))).)))))))......))....... ( -35.40) >DroEre_CAF1 11077 99 - 1 AAACAAAGCAGUACAAAGCAGUCGA-AUACACAGCUUGGUAGCGCGUUUACUGCUUGACAGUAGAUCACAUAGCAAGCAGU-GAUGCGAGCAGUUCAAGUC---- .......((..(((.((((.((...-....)).)))).))).((((((.((((((((...((.....))....))))))))-)))))).))..........---- ( -25.80) >DroYak_CAF1 9712 104 - 1 AAGCAAUGCAGUACAAAGCAGUCGAAAUCCCCUGCUUGAUAGCGUGUUUACUGCUUAAUGCGAGAUCAAUACGCAAGCAGU-CAUACAAGCAGGUCAAGCAGGUC ......(((........)))..........((((((((((.((.(((..((((((...((((.........))))))))))-...))).))..)))))))))).. ( -34.20) >consensus AAGCAAAGCAGUACAAAGCAGUCAAGAUAUACUGCUUUAUAUUGUGUUUACUGCUUGAUAUUAGAUCACUAAGCAAGCAGU_CAUGCAAGCAGUUCAAGCAGAUC .......((.....((((((((........))))))))....(((((.(((((((((...((((....)))).))))))))).))))).)).............. (-22.96 = -24.72 + 1.76)

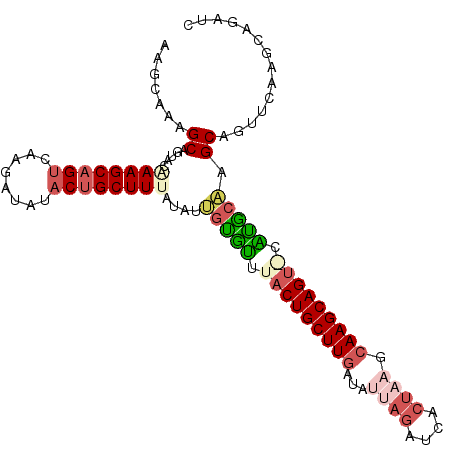
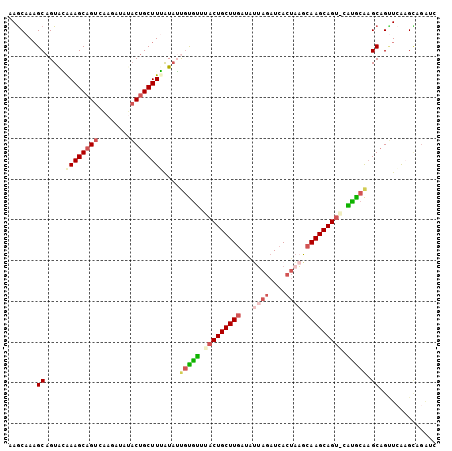
| Location | 2,733,969 – 2,734,064 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 72.62 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -10.55 |
| Energy contribution | -11.66 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2733969 95 - 27905053 ACUAUUACAGCGAACCUCGUGCAGCGAAGCAAAGCAGUAAAA--AGCAGUCUAGAUGUACUGCUUUAUAUUGUGUUUCCUGCUUGAUAUUAGAUCAC- .(((.(((((((.....)))((((.(((((...((((((.((--(((((((.....).)))))))).))))))))))))))).)).)).))).....- ( -29.30) >DroSec_CAF1 9909 95 - 1 ACUUUUACAGCGAACCUCGUGCAGCGAAGCAAAGCAGUGGGA--AGCAGUCAAGAUAUACUGCUUUAUAUUAUGUUUACUGCUUGAUAUUAGAUCAC- .((((....(((.....))).....))))..(((((((((((--((((((........)))))))).........)))))))))(((.....)))..- ( -21.10) >DroSim_CAF1 10640 95 - 1 ACUUUUACAGCGAACCUCGUGCUGCGAAGCAAAGCAGUAGGA--AGCAGUCAAGAUAUACUGCUUUAUUUUGUACUUACUGCUUGAUAUUAGAUCAC- ..(((..(((((.......)))))..)))..(((((((((((--((((((........))))))).........))))))))))(((.....)))..- ( -25.30) >DroEre_CAF1 11108 94 - 1 ACUUUUACAGCGAACCUCGUGCUGCGAAACAAAGCAGUACAA--AGCAGUCGA-AUACACAGCUUGGUAGCGCGUUUACUGCUUGACAGUAGAUCAC- .........(((.(((..(((((((........))))))).(--(((.((...-....)).)))))))..)))((((((((.....))))))))...- ( -27.90) >DroYak_CAF1 9747 95 - 1 ACUUUUACAGCGAGCCUCGUGCUGCGAAGCAAUGCAGUACAA--AGCAGUCGAAAUCCCCUGCUUGAUAGCGUGUUUACUGCUUAAUGCGAGAUCAA- ...........((..(((((((((((......)))))))).(--((((((.(((......((((....))))..)))))))))).....))).))..- ( -26.20) >DroPer_CAF1 28656 78 - 1 AGCUUUAAAGCGAACCUCGUUUC------CAAAGCAGUUCUUCAUGG---AGA-UAACACUGCUUC----------AACUGCUUGUCUGUUGAUCCAA .(((((.(((((.....))))).------.))))).........(((---(..-(((((..((...----------....)).....))))).)))). ( -16.80) >consensus ACUUUUACAGCGAACCUCGUGCUGCGAAGCAAAGCAGUACAA__AGCAGUCGAGAUACACUGCUUUAUAUUGUGUUUACUGCUUGAUAUUAGAUCAC_ .........(((.....)))...........((((((((.....((((((........))))))............)))))))).............. (-10.55 = -11.66 + 1.11)

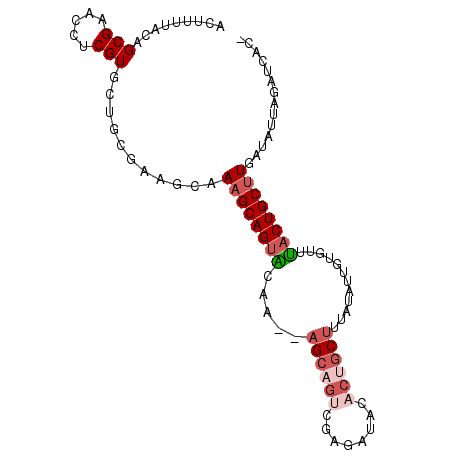
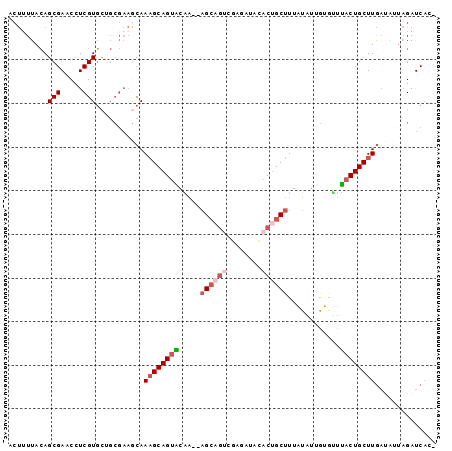
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:36 2006