| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,599,630 – 21,599,748 |
| Length | 118 |
| Max. P | 0.573782 |

| Location | 21,599,630 – 21,599,748 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.45 |
| Mean single sequence MFE | -36.34 |
| Consensus MFE | -24.85 |
| Energy contribution | -26.29 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21599630 118 + 27905053 UUUCAGAAUGCACGCAUCACAGAUGUUGCAGUGCCAGGACCUUG-GCG-GUACAUUCCUUUGGCAUAUUUUGCAAAAAUGCCACAGGUGCUCUUCACCUUUCAUUGGCUGCUGCAUUUGC .....(((((((.(((((...))))).((((((((((....)))-))(-((.....(((.((((((...........)))))).)))........)))........)))))))))))).. ( -35.42) >DroSec_CAF1 88523 120 + 1 NNNNNNNNNGCACGCAUCACAGGUGUUGCAGUGCCAGGACCGUGCGCGUGUACAUUCCUUUGGCAUAUUUUGCAAAAAUGCCACAGGUGCUCUUCACCUUUCAUCGGCUGCUGCAUUUCC .........(((.(((((...(((.((((((((((((((..((((....))))..)))..))))))....)))))....)))..(((((.....)))))......)).))))))...... ( -33.80) >DroSim_CAF1 84235 118 + 1 UUUUAGAAUGCACGCAUCACAGGUGUUGCAGUGCCAGGACCUUG-GCG-GUACAUUCCUUUGGCAUAUUUUGCAAAAAUGCCACAGGUGCUCCUCACCUUUCAUCGGCUGCUGCAUUUCC .....(((((((.(((.(..(((((..(((.((((((....)))-)))-.......(((.((((((...........)))))).))))))....))))).......).)))))))))).. ( -35.60) >DroEre_CAF1 79093 118 + 1 CUUCAGAAUGCACGCAUUACAGGUAUCGCAGUGCCAGGACCUUG-GCG-GCACUGUCCUCUGGCAUAUGUUGCAAUAGUGCCACAGAUGCUCCUCACCUUUCAUCGGCUGCUGCAUUUUA ....((((((((.(((.....((((((.....(((((....)))-))(-(((((((......(((.....))).))))))))...)))))).....((.......)).))))))))))). ( -38.20) >DroYak_CAF1 83393 118 + 1 UUUCAGAAUGCACGCAUUGCACGUCUGGCAGUGCCAGGACCUUG-GCG-GCACUGUCCUUUGGCAUAUGUUGCAAAAGUGCCACAGGUGCUCCUCACCUUCCAUCGGUUGCUGCACUUCC .....(((((((.(((..((((....(((((((((.........-..)-))))))))((.((((((...........)))))).)))))).....(((.......)))))))))).))). ( -38.70) >consensus UUUCAGAAUGCACGCAUCACAGGUGUUGCAGUGCCAGGACCUUG_GCG_GUACAUUCCUUUGGCAUAUUUUGCAAAAAUGCCACAGGUGCUCCUCACCUUUCAUCGGCUGCUGCAUUUCC .....(((((((.(((.........((((((((((((((................)))..))))))....)))))....(((..(((((.....)))))......))))))))))))).. (-24.85 = -26.29 + 1.44)

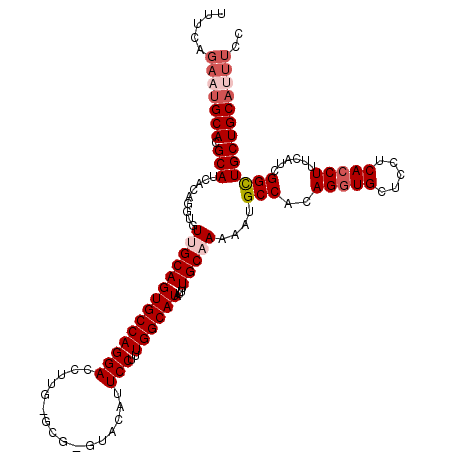
| Location | 21,599,630 – 21,599,748 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.45 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -28.43 |
| Energy contribution | -28.87 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21599630 118 - 27905053 GCAAAUGCAGCAGCCAAUGAAAGGUGAAGAGCACCUGUGGCAUUUUUGCAAAAUAUGCCAAAGGAAUGUAC-CGC-CAAGGUCCUGGCACUGCAACAUCUGUGAUGCGUGCAUUCUGAAA (((..(((((..((((.....(((((.....))))).))))....))))).....)))....(((((((((-.((-((......))))...(((.((....)).)))))))))))).... ( -37.10) >DroSec_CAF1 88523 120 - 1 GGAAAUGCAGCAGCCGAUGAAAGGUGAAGAGCACCUGUGGCAUUUUUGCAAAAUAUGCCAAAGGAAUGUACACGCGCACGGUCCUGGCACUGCAACACCUGUGAUGCGUGCNNNNNNNNN (((..(((.((..........(((((.....)))))((((((((((((((.....)))...)))))))).))))))))...)))..((((.(((.((....)).)))))))......... ( -33.00) >DroSim_CAF1 84235 118 - 1 GGAAAUGCAGCAGCCGAUGAAAGGUGAGGAGCACCUGUGGCAUUUUUGCAAAAUAUGCCAAAGGAAUGUAC-CGC-CAAGGUCCUGGCACUGCAACACCUGUGAUGCGUGCAUUCUAAAA ((((.(((((((..((.....(((((.((.((((((.((((((...........)))))).)))..))).)-)((-((......)))).......))))).)).))).)))))))).... ( -39.00) >DroEre_CAF1 79093 118 - 1 UAAAAUGCAGCAGCCGAUGAAAGGUGAGGAGCAUCUGUGGCACUAUUGCAACAUAUGCCAGAGGACAGUGC-CGC-CAAGGUCCUGGCACUGCGAUACCUGUAAUGCGUGCAUUCUGAAG ...(((((((((((((.....(((((.....))))).))))..(((((((.....(((((..((((..((.-...-))..))))))))).))))))).......))).))))))...... ( -37.10) >DroYak_CAF1 83393 118 - 1 GGAAGUGCAGCAACCGAUGGAAGGUGAGGAGCACCUGUGGCACUUUUGCAACAUAUGCCAAAGGACAGUGC-CGC-CAAGGUCCUGGCACUGCCAGACGUGCAAUGCGUGCAUUCUGAAA ((((.((((((((((.......))).....((((.(.((((.((((.(((.....))).))))...(((((-(((-....))...)))))))))).).))))..))).)))))))).... ( -41.90) >consensus GGAAAUGCAGCAGCCGAUGAAAGGUGAGGAGCACCUGUGGCAUUUUUGCAAAAUAUGCCAAAGGAAUGUAC_CGC_CAAGGUCCUGGCACUGCAACACCUGUGAUGCGUGCAUUCUGAAA ...(((((((((((((.....(((((.....))))).))))....(((((.....(((((..(((................)))))))).))))).........))).))))))...... (-28.43 = -28.87 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:01:01 2006