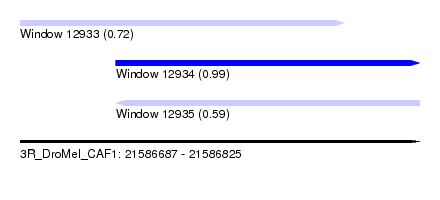
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,586,687 – 21,586,825 |
| Length | 138 |
| Max. P | 0.992917 |

| Location | 21,586,687 – 21,586,799 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 83.56 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -12.04 |
| Energy contribution | -11.92 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
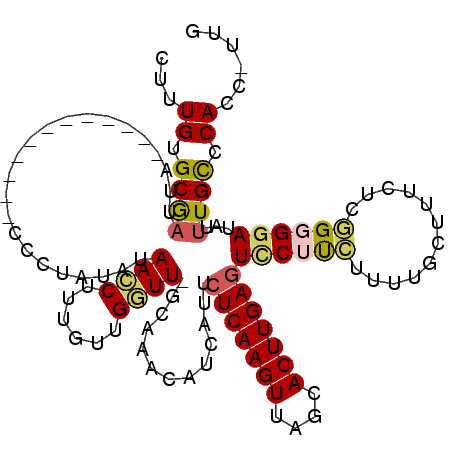
>3R_DroMel_CAF1 21586687 112 + 27905053 UUAAUUUAAAA-AUACGUUUUGUUAUUUUAUACUCAA-GUUGGGCAAAUAUCCCCUGAGAAAGAAAAAGAAGGACUCAAGUGCUAACUUGAGAAUGAUGUUUGCGAACCAACAA ........(((-(((........))))))........-((((((((((((((.(((..............))).(((((((....)))))))...)))))))))...))))).. ( -27.34) >DroSec_CAF1 75358 103 + 1 UUAAUUUAAAA-AUACUUUUUGUUAUUUUAUACUCAA-GGUGGGCAAAUAUCCCCCGAGAAAGCAAAAGAAGGACUCAAGUGCUAACUUGAGAAUG---------AACCAACAA ...........-.......(((((..(((((.(((((-((.(((........)))).....((((...((.....))...))))..)))))).)))---------))..))))) ( -20.00) >DroSim_CAF1 70617 103 + 1 UUAAUUUAAAA-AUACUUUUUGUUAUUUUAUACUCAA-GGUGGGCAAAUAUCCCCCGAGAAAGCAAAAGAAGGAUUCAAGUGCUAACUUGAGAAUG---------AACCAACAA ...........-.......(((((..(((((.(((((-(...((((...((((..(............)..)))).....))))..)))))).)))---------))..))))) ( -21.30) >DroEre_CAF1 65828 112 + 1 UUAACAUAUAA-AUAAUUUUUGUUAUUUUGUACUCAA-AGUGGGCAAAACUGCACAGAGAAGGCAAAAUGAGGACUCAAGUGCUAACUUGAGAAUGAUGUUUGCAAAUCAAAUA ..(((((....-.......((.(((((((((.(((..-.(..(......)..)...)))...))))))))).))(((((((....)))))))....)))))............. ( -25.50) >DroYak_CAF1 69897 114 + 1 UUAAUUUAUAAAAUACUUUUUGUUAUUUUGUACUCAAAAGUGGACAGAUGUCCACAGAGAAAGCAAAAUAAGAACUCAAGUGCUAACUUGAGAAUGAUGUUUGCAAAUCAAAGA ...................((.(((((((((.(((....((((((....)))))).)))...))))))))).))(((((((....)))))))..((((........)))).... ( -29.50) >consensus UUAAUUUAAAA_AUACUUUUUGUUAUUUUAUACUCAA_GGUGGGCAAAUAUCCCCAGAGAAAGCAAAAGAAGGACUCAAGUGCUAACUUGAGAAUGAUGUUUGC_AACCAACAA .................(((((((.((((..(((....)))(((......)))...)))).)))))))......(((((((....)))))))...................... (-12.04 = -11.92 + -0.12)

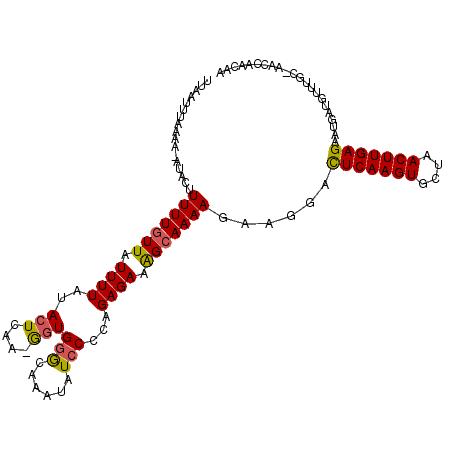
| Location | 21,586,720 – 21,586,825 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.12 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -13.84 |
| Energy contribution | -13.08 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.55 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21586720 105 + 27905053 CAA-GUUGGGCAAAUAUCCCCUGAGAAAGAAAAAGAAGGACUCAAGUGCUAACUUGAGAAUGAUGUUUGCGAACCAACAAAGGUUAUAUAGGG------------UAAUCGCACAAAG ...-((((((((((((((.(((..............))).(((((((....)))))))...)))))))))...)))))...((((((.....)------------)))))........ ( -30.54) >DroSec_CAF1 75391 96 + 1 CAA-GGUGGGCAAAUAUCCCCCGAGAAAGCAAAAGAAGGACUCAAGUGCUAACUUGAGAAUG---------AACCAACAAAGGUUAUAUAGGG------------UAAUCGCACAAAG ...-((.(((.......)))))......((..........(((((((....)))))))....---------..........((((((.....)------------)))))))...... ( -21.20) >DroSim_CAF1 70650 96 + 1 CAA-GGUGGGCAAAUAUCCCCCGAGAAAGCAAAAGAAGGAUUCAAGUGCUAACUUGAGAAUG---------AACCAACAAAGGUUAUAUAGGG------------UAAUCGCACAAAG ...-((.(((.......)))))......((..........(((((((....)))))))....---------..........((((((.....)------------)))))))...... ( -18.70) >DroEre_CAF1 65861 117 + 1 CAA-AGUGGGCAAAACUGCACAGAGAAGGCAAAAUGAGGACUCAAGUGCUAACUUGAGAAUGAUGUUUGCAAAUCAAAUAAGGUUAUAUAGGGUUUUCAGACAGAUAAUUGCACAAAA ...-.(..(......)..).........((((........(((((((....))))))).....((((((.(((((..(((......)))..))))).)))))).....))))...... ( -25.40) >DroYak_CAF1 69931 118 + 1 CAAAAGUGGACAGAUGUCCACAGAGAAAGCAAAAUAAGAACUCAAGUGCUAACUUGAGAAUGAUGUUUGCAAAUCAAAGAAGAUUAUAUAGGUGUUUUAAAAAGUUAAUUGCACAAAG .....((((((....)))))).......(((((.......(((((((....))))))).......))))).((((......))))......((((.((((....))))..)))).... ( -29.24) >consensus CAA_GGUGGGCAAAUAUCCCCAGAGAAAGCAAAAGAAGGACUCAAGUGCUAACUUGAGAAUGAUGUUUGC_AACCAACAAAGGUUAUAUAGGG____________UAAUCGCACAAAG .....(((.(..............................(((((((....))))))).............((((......))))........................).))).... (-13.84 = -13.08 + -0.76)

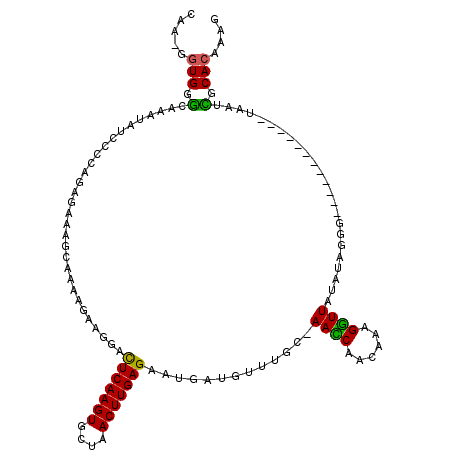

| Location | 21,586,720 – 21,586,825 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.12 |
| Mean single sequence MFE | -20.42 |
| Consensus MFE | -11.26 |
| Energy contribution | -11.42 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21586720 105 - 27905053 CUUUGUGCGAUUA------------CCCUAUAUAACCUUUGUUGGUUCGCAAACAUCAUUCUCAAGUUAGCACUUGAGUCCUUCUUUUUCUUUCUCAGGGGAUAUUUGCCCAAC-UUG ..(((.((((.((------------((((....((((......)))).............(((((((....)))))))...................)))).)).)))).))).-... ( -21.70) >DroSec_CAF1 75391 96 - 1 CUUUGUGCGAUUA------------CCCUAUAUAACCUUUGUUGGUU---------CAUUCUCAAGUUAGCACUUGAGUCCUUCUUUUGCUUUCUCGGGGGAUAUUUGCCCACC-UUG ...((.((((.((------------((((....((((......))))---------....(((((((....)))))))...................)))).)).)))).))..-... ( -20.90) >DroSim_CAF1 70650 96 - 1 CUUUGUGCGAUUA------------CCCUAUAUAACCUUUGUUGGUU---------CAUUCUCAAGUUAGCACUUGAAUCCUUCUUUUGCUUUCUCGGGGGAUAUUUGCCCACC-UUG ...((.((((.((------------((((....((((......))))---------......(.((..((((...((.....))...))))..)).))))).)).)))).))..-... ( -18.70) >DroEre_CAF1 65861 117 - 1 UUUUGUGCAAUUAUCUGUCUGAAAACCCUAUAUAACCUUAUUUGAUUUGCAAACAUCAUUCUCAAGUUAGCACUUGAGUCCUCAUUUUGCCUUCUCUGUGCAGUUUUGCCCACU-UUG ...((.((((....((((..((.......(((......)))..((...(((((.......(((((((....))))))).......)))))..))))...))))..)))).))..-... ( -18.44) >DroYak_CAF1 69931 118 - 1 CUUUGUGCAAUUAACUUUUUAAAACACCUAUAUAAUCUUCUUUGAUUUGCAAACAUCAUUCUCAAGUUAGCACUUGAGUUCUUAUUUUGCUUUCUCUGUGGACAUCUGUCCACUUUUG .................................((((......)))).(((((.......(((((((....))))))).......))))).......((((((....))))))..... ( -22.34) >consensus CUUUGUGCGAUUA____________CCCUAUAUAACCUUUGUUGGUU_GCAAACAUCAUUCUCAAGUUAGCACUUGAGUCCUUCUUUUGCUUUCUCGGGGGAUAUUUGCCCACC_UUG ...((.((((.......................((((......)))).............(((((((....)))))))((((((............))))))...)))).))...... (-11.26 = -11.42 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:57 2006