
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,575,671 – 21,575,805 |
| Length | 134 |
| Max. P | 0.999923 |

| Location | 21,575,671 – 21,575,770 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 74.19 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -16.10 |
| Energy contribution | -16.66 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.58 |
| SVM decision value | 3.63 |
| SVM RNA-class probability | 0.999466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21575671 99 + 27905053 GUGAGUAUAUGUAUGUAUGUGUGGCCACGCCCACACGCC-CUGUUUAUAAACACACAC----ACUGCCACACACAUAUUUAUUUACACGCACUUGAGCACGUGC (((((((...((((((..(((((((...((......)).-.((((....)))).....----...))))))))))))).)))))))..((((........)))) ( -26.90) >DroSec_CAF1 63938 90 + 1 GUGAGUAUAUGUACGUAUGUGUGGCCACGCCCACACGUGUGUGUUUAUGAAC--------------ACACACACAUAUUUAUUUACUCGCACUUGAGCACGUGC (((((((...(((.((((((((((......))))..(((((((((....)))--------------)))))))))))).))).))))))).............. ( -33.40) >DroSim_CAF1 59051 103 + 1 GUGAGUAUAUGUACGUAUGUGUGGCCACGCCCACACGCC-CUGUUUAUAAACAUAUACGACCACACACACACACAUAUUUAUUUACACGCACUUGAGCACGUGC ((((((((((((..((.(((((((.(..((......)).-.((((....)))).....).))))))).))..)))))..)))))))..((((........)))) ( -25.40) >DroYak_CAF1 58102 85 + 1 GUGAGUAUAU--------GUGUGGCCACGCCCACACGCC-CCGUUUAUAAACACACA----------CACACACAUAUGUAUUUACACGCACUUGAGCACGUGC ((((((((((--------(((((.....((......)).-..(((....))).....----------....)))))))))))))))..((((........)))) ( -22.60) >DroAna_CAF1 45707 97 + 1 AUGAAUGCCUGUGUG--UGUGUGGAUGCGACCACACCCA-CCGCUCU-A---CUGUGCCCCCAAACACACAUACAUAGUUAUUUACACGCACUUGAGCACGUGC .((((((.(((((((--((((((..((.(..((((....-.......-.---.))))..).))..))))))))))))).))))))...((((........)))) ( -30.62) >consensus GUGAGUAUAUGUACGUAUGUGUGGCCACGCCCACACGCC_CUGUUUAUAAACACACAC____A___ACACACACAUAUUUAUUUACACGCACUUGAGCACGUGC (((((((.((((......((((((......))))))......(((....)))......................)))).)))))))..((((........)))) (-16.10 = -16.66 + 0.56)


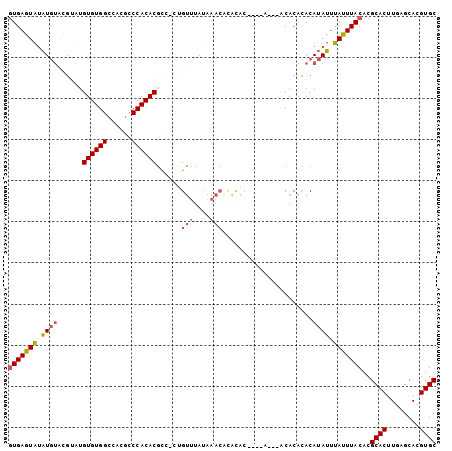
| Location | 21,575,671 – 21,575,770 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 74.19 |
| Mean single sequence MFE | -36.56 |
| Consensus MFE | -21.56 |
| Energy contribution | -22.72 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.80 |
| Structure conservation index | 0.59 |
| SVM decision value | 4.57 |
| SVM RNA-class probability | 0.999923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21575671 99 - 27905053 GCACGUGCUCAAGUGCGUGUAAAUAAAUAUGUGUGUGGCAGU----GUGUGUGUUUAUAAACAG-GGCGUGUGGGCGUGGCCACACAUACAUACAUAUACUCAC (((((..(....)..)))))......(((((((((((((..(----((.(((....))).))).-.))((((((......)))))).)))))))))))...... ( -36.70) >DroSec_CAF1 63938 90 - 1 GCACGUGCUCAAGUGCGAGUAAAUAAAUAUGUGUGUGU--------------GUUCAUAAACACACACGUGUGGGCGUGGCCACACAUACGUACAUAUACUCAC ((((........))))(((((.((...(((((((((((--------------(..(((...((((....))))...)))..)))))))))))).)).))))).. ( -36.40) >DroSim_CAF1 59051 103 - 1 GCACGUGCUCAAGUGCGUGUAAAUAAAUAUGUGUGUGUGUGUGGUCGUAUAUGUUUAUAAACAG-GGCGUGUGGGCGUGGCCACACAUACGUACAUAUACUCAC (((((..(....)..)))))......((((((((((((((((((((((((((((((.......)-))))))))....)))))))))))))))))))))...... ( -43.50) >DroYak_CAF1 58102 85 - 1 GCACGUGCUCAAGUGCGUGUAAAUACAUAUGUGUGUG----------UGUGUGUUUAUAAACGG-GGCGUGUGGGCGUGGCCACAC--------AUAUACUCAC (((((..(....)..)))))((((((((((......)----------)))))))))......((-(..((((((((...))).)))--------))...))).. ( -30.40) >DroAna_CAF1 45707 97 - 1 GCACGUGCUCAAGUGCGUGUAAAUAACUAUGUAUGUGUGUUUGGGGGCACAG---U-AGAGCGG-UGGGUGUGGUCGCAUCCACACA--CACACAGGCAUUCAU (((((..(....)..)))))........((((.(((((((......((....---.-...)).(-(((((((....))))))))..)--)))))).)))).... ( -35.80) >consensus GCACGUGCUCAAGUGCGUGUAAAUAAAUAUGUGUGUGU___U____GUAUAUGUUUAUAAACAG_GGCGUGUGGGCGUGGCCACACAUACAUACAUAUACUCAC (((((..(....)..))))).........((((((((...............(((....)))......((((((......)))))).....))))))))..... (-21.56 = -22.72 + 1.16)


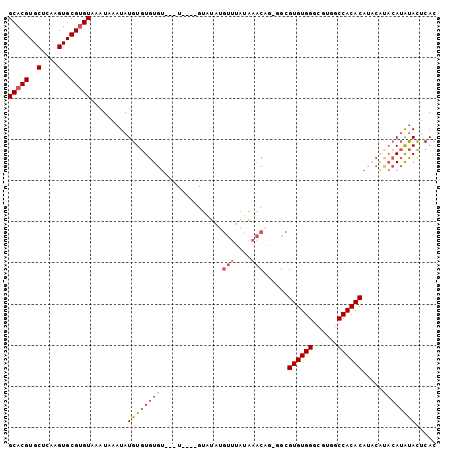
| Location | 21,575,702 – 21,575,805 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.56 |
| Mean single sequence MFE | -18.42 |
| Consensus MFE | -8.87 |
| Energy contribution | -9.37 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21575702 103 + 27905053 CACACGCC-CUGUUUAUAAACACACAC----ACUGCCACACACAUAUUUAUUUACACGCACUUGAGCACGUGCUGAGCAGGGAAAAUAC--CCCACAAGCACAAACAAAA ......((-(((((((...........----..((.....))...............((((........))))))))))))).......--................... ( -15.80) >DroSec_CAF1 63969 94 + 1 CACACGUGUGUGUUUAUGAAC--------------ACACACACAUAUUUAUUUACUCGCACUUGAGCACGUGCUGAGCAGGGAAAAUAA--CCCACAAGCACAAACAAAA .....(((((((((....)))--------------))))))................((......))..(((((.....(((.......--)))...)))))........ ( -25.10) >DroSim_CAF1 59082 107 + 1 CACACGCC-CUGUUUAUAAACAUAUACGACCACACACACACACAUAUUUAUUUACACGCACUUGAGCACGUGCUGAGCAGGGAAAAUAC--CCCACAAGCACAAACAAAA ......((-(((((((....(......).............................((((........))))))))))))).......--................... ( -15.80) >DroEre_CAF1 54326 95 + 1 CACACGCC-CCCUUCAUAAACACACA------------CACACAUGUGUAUUUACACGCACUUGAGCACGUGCUGAGCAGGGAAAAUUC--CCCACAAGCACAAACAAAA ........-.......((((.(((((------------......))))).))))...((......))..(((((.....((((....))--))....)))))........ ( -19.50) >DroYak_CAF1 58125 97 + 1 CACACGCC-CCGUUUAUAAACACACA----------CACACACAUAUGUAUUUACACGCACUUGAGCACGUGCUGAGGAGGGAAAAUAC--CCCACAAGCACAAACAAAA ........-..((((...........----------......((..(((........)))..)).....(((((...(.(((......)--)))...))))))))).... ( -16.60) >DroAna_CAF1 45736 103 + 1 CACACCCA-CCGCUCU-A---CUGUGCCCCCAAACACACAUACAUAGUUAUUUACACGCACUUGAGCACGUGCUGAGCAGGGAGAAUAUAUACCAUAA--ACAAACAAAA ....(((.-..((((.-(---(((((................)))))).........((((........)))).)))).)))................--.......... ( -17.69) >consensus CACACGCC_CCGUUUAUAAACACACA_________ACACACACAUAUUUAUUUACACGCACUUGAGCACGUGCUGAGCAGGGAAAAUAC__CCCACAAGCACAAACAAAA .........................................................((......))..(((((.....(((.........)))...)))))........ ( -8.87 = -9.37 + 0.50)


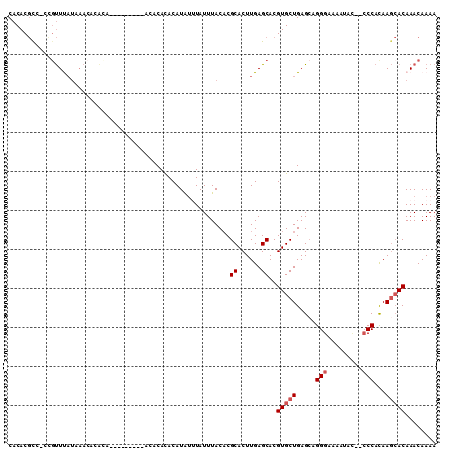
| Location | 21,575,702 – 21,575,805 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.56 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -14.62 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21575702 103 - 27905053 UUUUGUUUGUGCUUGUGGG--GUAUUUUCCCUGCUCAGCACGUGCUCAAGUGCGUGUAAAUAAAUAUGUGUGUGGCAGU----GUGUGUGUUUAUAAACAG-GGCGUGUG .((((((((((...(..((--(......)))..)...(((((..(((..(..(((((......)))))..)...).)).----.)))))...)))))))))-)....... ( -30.90) >DroSec_CAF1 63969 94 - 1 UUUUGUUUGUGCUUGUGGG--UUAUUUUCCCUGCUCAGCACGUGCUCAAGUGCGAGUAAAUAAAUAUGUGUGUGU--------------GUUCAUAAACACACACGUGUG ..(((((((((((.(..((--........))..)..)))))..((((......))))))))))((((((((((((--------------........)))))))))))). ( -33.60) >DroSim_CAF1 59082 107 - 1 UUUUGUUUGUGCUUGUGGG--GUAUUUUCCCUGCUCAGCACGUGCUCAAGUGCGUGUAAAUAAAUAUGUGUGUGUGUGUGGUCGUAUAUGUUUAUAAACAG-GGCGUGUG ........(..(....(((--(......))))((((.(((((..(....)..)))))..(((((((((((((..........))))))))))))).....)-))))..). ( -29.40) >DroEre_CAF1 54326 95 - 1 UUUUGUUUGUGCUUGUGGG--GAAUUUUCCCUGCUCAGCACGUGCUCAAGUGCGUGUAAAUACACAUGUGUG------------UGUGUGUUUAUGAAGGG-GGCGUGUG ........(((((.(..((--((....))))..)..)))))..((((.......(((((((((((((....)------------))))))))))))....)-)))..... ( -34.90) >DroYak_CAF1 58125 97 - 1 UUUUGUUUGUGCUUGUGGG--GUAUUUUCCCUCCUCAGCACGUGCUCAAGUGCGUGUAAAUACAUAUGUGUGUG----------UGUGUGUUUAUAAACGG-GGCGUGUG ........(((((.(..((--(......)))..)..)))))..((((..((...(((((((((((((......)----------)))))))))))).)).)-)))..... ( -31.90) >DroAna_CAF1 45736 103 - 1 UUUUGUUUGU--UUAUGGUAUAUAUUCUCCCUGCUCAGCACGUGCUCAAGUGCGUGUAAAUAACUAUGUAUGUGUGUUUGGGGGCACAG---U-AGAGCGG-UGGGUGUG ..........--........(((((((...((((((.(((((..(....)..)))))...............((((((....)))))).---.-.))))))-.))))))) ( -28.90) >consensus UUUUGUUUGUGCUUGUGGG__GUAUUUUCCCUGCUCAGCACGUGCUCAAGUGCGUGUAAAUAAAUAUGUGUGUGU_________UGUGUGUUUAUAAACAG_GGCGUGUG ...((((((((.....(((.........)))......(((((..(....)..)))))...................................)))))))).......... (-14.62 = -14.90 + 0.28)


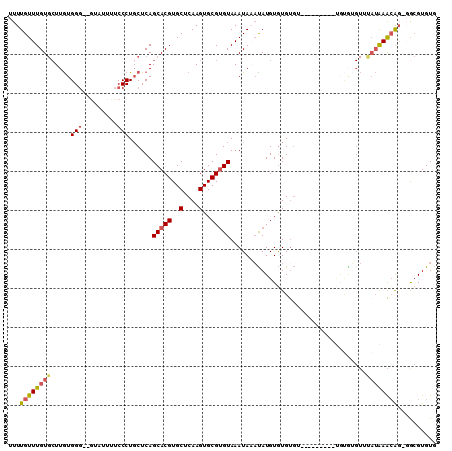
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:54 2006