
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,568,773 – 21,568,905 |
| Length | 132 |
| Max. P | 0.990244 |

| Location | 21,568,773 – 21,568,884 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.64 |
| Mean single sequence MFE | -26.81 |
| Consensus MFE | -17.92 |
| Energy contribution | -18.09 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
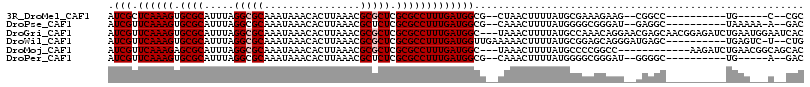
>3R_DroMel_CAF1 21568773 111 + 27905053 CUCC---AUUCUCCACAGCAGCGAAGGAAUUUUCAAUCAAAUCGCUCAAAGUGCGCAUUUAGGCGCAAAUAAACACUUAAACGCGCUCGCGCCUUUGAUGGCG--CUAACUUUUAU---- ....---..........(.(((((..((........))...))))))(((((((((.((((((.(........).)))))).))))..(((((......))))--)..)))))...---- ( -29.50) >DroVir_CAF1 45535 106 + 1 -------UUCCCCGGCCACUGCAAAAGAAUUUUCAAUCAAAUCGUUCAAAGUGCGCAUUUAGGCGCAAAUAAACACUUAAACGCGCUCGCGCCUUUGAUGGG---UAAACUUUUAU---- -------...((((((....)).......................((((((.((((.....(((((................))))).))))))))))))))---...........---- ( -23.69) >DroWil_CAF1 38344 113 + 1 UGCC---AUUCGUCGUCGUUCGCAAAGAAUUUCCAAUCAAAUCGUUCAAAGUGCGCAUUUAGGCGCAAAUAAACACUUAAACGCGCUCGCGCCUUUGAUGGUUGAAAAACUUUUAU---- (((.---...((....))...)))............((((.((((.(((((.((((.....(((((................))))).)))))))))))))))))...........---- ( -24.39) >DroMoj_CAF1 43785 109 + 1 -UCG---UUCCCCUGCCAAUGCAAAAGAAUUUUCAAUCAAAUCGUUCAAAGAGCGCAUUUAGGCGCAAAUAAACACUUAAACGCGCUCGCGCCUUUGAUGGC---UAAACUUUUAU---- -..(---(((...(((....)))...))))....................((((((.((((((.(........).)))))).))))))..(((......)))---...........---- ( -23.60) >DroAna_CAF1 39236 118 + 1 CGCCACCCACCGGCGGAGAUGCGAAGGAAUUUUCAAUCAAAUCGCUCAAAGUGCGCAUUUAGGCGCAAAUAAACACUUAAACGCGCUCGCGCCUUUGAUGGCG--CAAACUUUUAUACGA ((((.......))))..((.((((..((........))...))))))(((((((((.((((((.(........).)))))).))))..(((((......))))--)..)))))....... ( -36.00) >DroPer_CAF1 52728 106 + 1 C--------CUCUCAGAGAUCCAAAAGAAUUUUCAAUCAAAUCGUUCAAAGUGCGCAUUUAGGCGCAAAUAAACACUUAAACGCUCUCGCGCCUUUGAUGGCG--CAAACUUUUAU---- .--------......((((.......((....)).........(((.....(((((......)))))....)))..........))))(((((......))))--)..........---- ( -23.70) >consensus C_CC___AUCCCCCGCAGAUGCAAAAGAAUUUUCAAUCAAAUCGUUCAAAGUGCGCAUUUAGGCGCAAAUAAACACUUAAACGCGCUCGCGCCUUUGAUGGCG__CAAACUUUUAU____ .........................................(((.((((((.((((.....(((((................))))).)))))))))))))................... (-17.92 = -18.09 + 0.17)



| Location | 21,568,773 – 21,568,884 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.64 |
| Mean single sequence MFE | -35.77 |
| Consensus MFE | -23.02 |
| Energy contribution | -24.05 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21568773 111 - 27905053 ----AUAAAAGUUAG--CGCCAUCAAAGGCGCGAGCGCGUUUAAGUGUUUAUUUGCGCCUAAAUGCGCACUUUGAGCGAUUUGAUUGAAAAUUCCUUCGCUGCUGUGGAGAAU---GGAG ----...(((((..(--((((......)))))..(((((((((.((((......)))).)))))))))))))).(((((...((.(....).))..))))).((((.....))---)).. ( -37.50) >DroVir_CAF1 45535 106 - 1 ----AUAAAAGUUUA---CCCAUCAAAGGCGCGAGCGCGUUUAAGUGUUUAUUUGCGCCUAAAUGCGCACUUUGAACGAUUUGAUUGAAAAUUCUUUUGCAGUGGCCGGGGAA------- ----...........---(((.....((((((((((((......))))....))))))))....((.((((..(((.(((((......))))).)))...)))))).)))...------- ( -30.00) >DroWil_CAF1 38344 113 - 1 ----AUAAAAGUUUUUCAACCAUCAAAGGCGCGAGCGCGUUUAAGUGUUUAUUUGCGCCUAAAUGCGCACUUUGAACGAUUUGAUUGGAAAUUCUUUGCGAACGACGACGAAU---GGCA ----......((..(((.....((((((((....))(((((((.((((......)))).)))))))...)))))).((.((((...((.....))...)))))).....))).---.)). ( -30.50) >DroMoj_CAF1 43785 109 - 1 ----AUAAAAGUUUA---GCCAUCAAAGGCGCGAGCGCGUUUAAGUGUUUAUUUGCGCCUAAAUGCGCUCUUUGAACGAUUUGAUUGAAAAUUCUUUUGCAUUGGCAGGGGAA---CGA- ----......(((((---(((......)))..(((((((((((.((((......)))).)))))))))))..)))))..............(((((((((....)))))))))---...- ( -38.30) >DroAna_CAF1 39236 118 - 1 UCGUAUAAAAGUUUG--CGCCAUCAAAGGCGCGAGCGCGUUUAAGUGUUUAUUUGCGCCUAAAUGCGCACUUUGAGCGAUUUGAUUGAAAAUUCCUUCGCAUCUCCGCCGGUGGGUGGCG ..(((........))--)((((((...((((.(((((((((((.((((......)))).))))))))).......((((...((.(....).))..))))...))))))....)))))). ( -45.00) >DroPer_CAF1 52728 106 - 1 ----AUAAAAGUUUG--CGCCAUCAAAGGCGCGAGAGCGUUUAAGUGUUUAUUUGCGCCUAAAUGCGCACUUUGAACGAUUUGAUUGAAAAUUCUUUUGGAUCUCUGAGAG--------G ----.......((((--((((......)))))))).(((((((.((((......)))).)))))))...((((.(..((((..(..((....))..)..))))..).))))--------. ( -33.30) >consensus ____AUAAAAGUUUG__CGCCAUCAAAGGCGCGAGCGCGUUUAAGUGUUUAUUUGCGCCUAAAUGCGCACUUUGAACGAUUUGAUUGAAAAUUCUUUUGCAUCGGCGGGGGAA___GG_G ......................((((((((....))(((((((.((((......)))).)))))))...)))))).................((((((((....))))))))........ (-23.02 = -24.05 + 1.03)



| Location | 21,568,810 – 21,568,905 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.01 |
| Mean single sequence MFE | -31.37 |
| Consensus MFE | -16.74 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21568810 95 + 27905053 AUCGCUCAAAGUGCGCAUUUAGGCGCAAAUAAACACUUAAACGCGCUCGCGCCUUUGAUGGCG--CUAACUUUUAUGCGAAAGAAG--CGGCC----------UG-----C--CGC .((((..(((((((((.((((((.(........).)))))).))))..(((((......))))--)..)))))...)))).....(--(((..----------..-----)--))) ( -36.20) >DroPse_CAF1 52468 99 + 1 AUCGUUCAAAGUGCGCAUUUAGGCGCAAAUAAACACUUAAACGCUCUCGCGCCUUUGAUGGCG--CAAACUUUUAUGGGGCGGGAU--GAGGC----------UAAAAA-A--GAC .((((((....(((((......)))))..............(((((..(((((......))))--)...(......))))))))))--))...----------......-.--... ( -30.80) >DroGri_CAF1 43467 113 + 1 AUCGUUCAAAGUGCGCAUUUAGGCGCAAAUAAACACUUAAACGCGCUCGCGCCUUUGAUGGC---UAAACUUUUAUGCCAAACAGGAACGAGCAACGGAGAUCUGAAUGGAAUCAC .(((((((...(((((......))))).................(((((..(((....((((---...........))))...)))..)))))..........)))))))...... ( -31.80) >DroWil_CAF1 38381 103 + 1 AUCGUUCAAAGUGCGCAUUUAGGCGCAAAUAAACACUUAAACGCGCUCGCGCCUUUGAUGGUUGAAAAACUUUUAUGCGGAGCAGGGAUGAGC----------UGAGUC-U--CUG ...(((.....(((((......)))))....)))..........((((.(((...(((.((((....)))).))).)))))))((((((....----------...)))-)--)). ( -29.70) >DroMoj_CAF1 43821 101 + 1 AUCGUUCAAAGAGCGCAUUUAGGCGCAAAUAAACACUUAAACGCGCUCGCGCCUUUGAUGGC---UAAACUUUUAUGCCCCGGCC------------AAGAUCUGAACGGCAGCAC .(((((((..((((((.((((((.(........).)))))).))))))(.(((......(((---...........)))..))))------------......)))))))...... ( -29.80) >DroPer_CAF1 52760 95 + 1 AUCGUUCAAAGUGCGCAUUUAGGCGCAAAUAAACACUUAAACGCUCUCGCGCCUUUGAUGGCG--CAAACUUUUAUGGGGCGGGAU--GGGGC----------UG-----A--GAC .((((((....(((((......)))))..............(((((..(((((......))))--)...(......))))))))))--))...----------..-----.--... ( -29.90) >consensus AUCGUUCAAAGUGCGCAUUUAGGCGCAAAUAAACACUUAAACGCGCUCGCGCCUUUGAUGGCG__CAAACUUUUAUGCGGAGGAAG__CGGGC__________UGAA___A__CAC .(((.((((((.((((.....(((((................))))).)))))))))))))....................................................... (-16.74 = -17.07 + 0.33)

| Location | 21,568,810 – 21,568,905 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.01 |
| Mean single sequence MFE | -33.47 |
| Consensus MFE | -20.50 |
| Energy contribution | -21.03 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
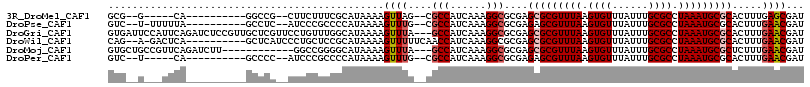
>3R_DroMel_CAF1 21568810 95 - 27905053 GCG--G-----CA----------GGCCG--CUUCUUUCGCAUAAAAGUUAG--CGCCAUCAAAGGCGCGAGCGCGUUUAAGUGUUUAUUUGCGCCUAAAUGCGCACUUUGAGCGAU (((--(-----..----------..)))--).....((((...(((((..(--((((......)))))..(((((((((.((((......)))).))))))))))))))..)))). ( -43.30) >DroPse_CAF1 52468 99 - 1 GUC--U-UUUUUA----------GCCUC--AUCCCGCCCCAUAAAAGUUUG--CGCCAUCAAAGGCGCGAGAGCGUUUAAGUGUUUAUUUGCGCCUAAAUGCGCACUUUGAACGAU ...--.-......----------...((--(....((..........((((--((((......)))))))).(((((((.((((......)))).)))))))))....)))..... ( -26.80) >DroGri_CAF1 43467 113 - 1 GUGAUUCCAUUCAGAUCUCCGUUGCUCGUUCCUGUUUGGCAUAAAAGUUUA---GCCAUCAAAGGCGCGAGCGCGUUUAAGUGUUUAUUUGCGCCUAAAUGCGCACUUUGAACGAU .(((......)))......((((((((((.(((...((((.(((....)))---))))....))).))))))(((((((.((((......)))).)))))))........)))).. ( -36.10) >DroWil_CAF1 38381 103 - 1 CAG--A-GACUCA----------GCUCAUCCCUGCUCCGCAUAAAAGUUUUUCAACCAUCAAAGGCGCGAGCGCGUUUAAGUGUUUAUUUGCGCCUAAAUGCGCACUUUGAACGAU (((--(-(....(----------((........))).(((((....(((....)))......((((((((((((......))))....))))))))..)))))..)))))...... ( -26.60) >DroMoj_CAF1 43821 101 - 1 GUGCUGCCGUUCAGAUCUU------------GGCCGGGGCAUAAAAGUUUA---GCCAUCAAAGGCGCGAGCGCGUUUAAGUGUUUAUUUGCGCCUAAAUGCGCUCUUUGAACGAU (((((.(((..(((...))------------)..))))))))....(((((---(((......)))..(((((((((((.((((......)))).)))))))))))..)))))... ( -40.30) >DroPer_CAF1 52760 95 - 1 GUC--U-----CA----------GCCCC--AUCCCGCCCCAUAAAAGUUUG--CGCCAUCAAAGGCGCGAGAGCGUUUAAGUGUUUAUUUGCGCCUAAAUGCGCACUUUGAACGAU .((--(-----(.----------((...--.....)).............(--((((......))))))))).(((((((((((.((((((....)))))).)))).))))))).. ( -27.70) >consensus GUG__U___UUCA__________GCCCC__CUUCCGCCGCAUAAAAGUUUG__CGCCAUCAAAGGCGCGAGCGCGUUUAAGUGUUUAUUUGCGCCUAAAUGCGCACUUUGAACGAU ..............................................((((....(((......)))....(((((((((.((((......)))).))))))))).....))))... (-20.50 = -21.03 + 0.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:49 2006