| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,732,841 – 2,732,949 |
| Length | 108 |
| Max. P | 0.919992 |

| Location | 2,732,841 – 2,732,949 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.46 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -17.99 |
| Energy contribution | -17.55 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2732841 108 + 27905053 AAUUGAUGGCAUCCAUUGCACAUAACGUGUAACUAAUGGCGCUCAUCAAUUGUGCUUUACAUCUCAAUCUGGCUUUGGCGAAA-AAA-U--------UCAGCCGGCUACA--AACAAAAA ((((((((((..(((((((((.....)))))....)))).)).))))))))..................(((((..((((((.-...-)--------)).))))))))..--........ ( -25.80) >DroVir_CAF1 48023 109 + 1 AAUUGAUGGCAUCCAUUGCACAUAACGUGUAACUAAUGGCGCUCAUCAAUUCUGCUUUACAUCUCAAUCUGGCUAAGGCAGAA-AAA-UAAAAGAGCACAAGCGACAGCA--A------- ..((((((((..(((((((((.....)))))....)))).)).))))))((((((((((..((.......)).))))))))))-...-.............((....)).--.------- ( -27.50) >DroPse_CAF1 28441 101 + 1 AAUUGAUGGCAUCCAUUGCACAUAACGUGUAACUAAUGGCGCUCAUCAAUUCUGCUUUACAUCUCAAUCUGGCUUUGGUGCAA-ACA-A--A-----AAAAC----------AGGAAAAA ((((((((((..(((((((((.....)))))....)))).)).))))))))................((((..((((......-.))-)--)-----....)----------)))..... ( -20.60) >DroMoj_CAF1 49651 104 + 1 AAUUGAUGGCAUCCAUUGCACAUAACGUGUAACUAAUGGCGCUCAUCAAUUCUGCUUUACAUCUCAAUCUGGCCAAGGCAGCG-AAA-UAAAAGA-----AGAGCGAGCA--A------- ((((((((((..(((((((((.....)))))....)))).)).)))))))).(((((..............((....)).((.-...-.......-----...)))))))--.------- ( -24.14) >DroAna_CAF1 8665 111 + 1 AAUUGAUGGCAUCCAUUGCACAUAACGUGUAACUAAUGGCGCUCAUCAAUUCUGCUUUACAUCUCAAUCUGGCUUUGGCGAAAAAAA-U--------ACAUCCAGCAGGAACAACAAAAA ..((((((((..(((((((((.....)))))....)))).)).))))))(((((((.............(.((....)).)......-.--------......))))))).......... ( -24.07) >DroPer_CAF1 27492 102 + 1 AAUUGAUGGCAUCCAUUGCACAUAACGUGUAACUAAUGGCGCUCAUCAAUUCUGCUUUACAUCUCAAUCUGGCUUUGGUGCAA-AAAAA--A-----AAAAC----------AGGAAAAA ((((((((((..(((((((((.....)))))....)))).)).))))))))................((((..(((..(....-..)..--)-----))..)----------)))..... ( -20.10) >consensus AAUUGAUGGCAUCCAUUGCACAUAACGUGUAACUAAUGGCGCUCAUCAAUUCUGCUUUACAUCUCAAUCUGGCUUUGGCGAAA_AAA_U__A_____ACAACCG_CAGCA__A__AAAAA ((((((((((..(((((((((.....)))))....)))).)).)))))))).((((...((........)).....))))........................................ (-17.99 = -17.55 + -0.44)

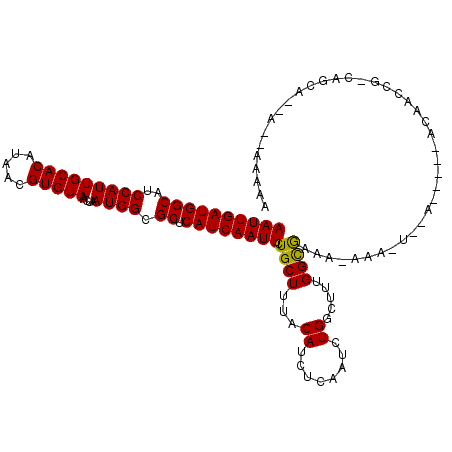
| Location | 2,732,841 – 2,732,949 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.46 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -18.15 |
| Energy contribution | -18.15 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2732841 108 - 27905053 UUUUUGUU--UGUAGCCGGCUGA--------A-UUU-UUUCGCCAAAGCCAGAUUGAGAUGUAAAGCACAAUUGAUGAGCGCCAUUAGUUACACGUUAUGUGCAAUGGAUGCCAUCAAUU ((((.(((--((..((.(((.((--------(-...-.))))))...))))))).))))..........((((((((.((((((((.((.(((.....)))))))))).))))))))))) ( -26.60) >DroVir_CAF1 48023 109 - 1 -------U--UGCUGUCGCUUGUGCUCUUUUA-UUU-UUCUGCCUUAGCCAGAUUGAGAUGUAAAGCAGAAUUGAUGAGCGCCAUUAGUUACACGUUAUGUGCAAUGGAUGCCAUCAAUU -------.--.......((((.(((((((...-...-.(((((....).))))..)))).)))))))..((((((((.((((((((.((.(((.....)))))))))).))))))))))) ( -27.50) >DroPse_CAF1 28441 101 - 1 UUUUUCCU----------GUUUU-----U--U-UGU-UUGCACCAAAGCCAGAUUGAGAUGUAAAGCAGAAUUGAUGAGCGCCAUUAGUUACACGUUAUGUGCAAUGGAUGCCAUCAAUU ......((----------(((((-----.--.-.((-((((......).)))))........)))))))((((((((.((((((((.((.(((.....)))))))))).))))))))))) ( -24.20) >DroMoj_CAF1 49651 104 - 1 -------U--UGCUCGCUCU-----UCUUUUA-UUU-CGCUGCCUUGGCCAGAUUGAGAUGUAAAGCAGAAUUGAUGAGCGCCAUUAGUUACACGUUAUGUGCAAUGGAUGCCAUCAAUU -------(--((((.((...-----((((..(-(((-.(((.....))).)))).)))).))..)))))((((((((.((((((((.((.(((.....)))))))))).))))))))))) ( -28.50) >DroAna_CAF1 8665 111 - 1 UUUUUGUUGUUCCUGCUGGAUGU--------A-UUUUUUUCGCCAAAGCCAGAUUGAGAUGUAAAGCAGAAUUGAUGAGCGCCAUUAGUUACACGUUAUGUGCAAUGGAUGCCAUCAAUU ............(((((..((((--------.-((..(((.((....)).)))..)).))))..)))))((((((((.((((((((.((.(((.....)))))))))).))))))))))) ( -29.00) >DroPer_CAF1 27492 102 - 1 UUUUUCCU----------GUUUU-----U--UUUUU-UUGCACCAAAGCCAGAUUGAGAUGUAAAGCAGAAUUGAUGAGCGCCAUUAGUUACACGUUAUGUGCAAUGGAUGCCAUCAAUU ......((----------(((((-----(--((..(-((((......).))))..))))....))))))((((((((.((((((((.((.(((.....)))))))))).))))))))))) ( -24.80) >consensus UUUUU__U__UGCUG_CGGUUGU_____U__A_UUU_UUCCGCCAAAGCCAGAUUGAGAUGUAAAGCAGAAUUGAUGAGCGCCAUUAGUUACACGUUAUGUGCAAUGGAUGCCAUCAAUU ...............................................((...((......))...))..((((((((.((((((((.((.(((.....)))))))))).))))))))))) (-18.15 = -18.15 + 0.00)

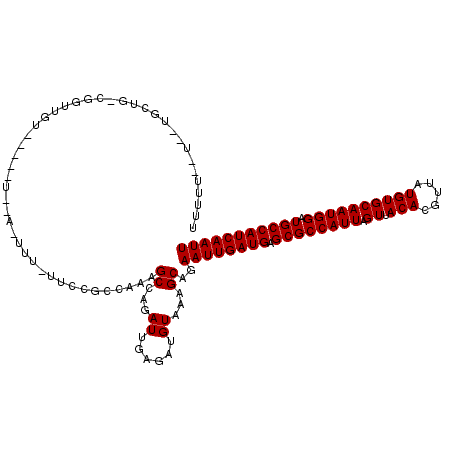
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:33 2006