
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,510,095 – 21,510,242 |
| Length | 147 |
| Max. P | 0.739830 |

| Location | 21,510,095 – 21,510,215 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.28 |
| Mean single sequence MFE | -49.17 |
| Consensus MFE | -35.74 |
| Energy contribution | -35.47 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
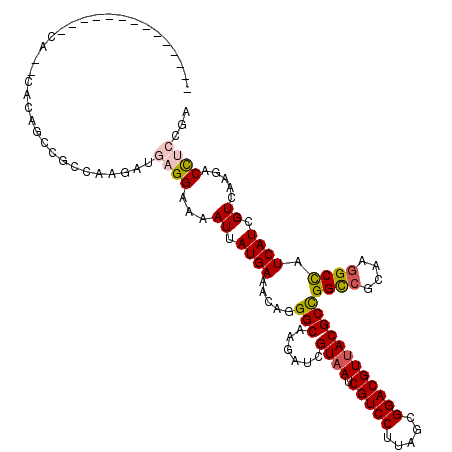
>3R_DroMel_CAF1 21510095 120 - 27905053 AUCAUGAAGCAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGUCGCAAGGCCAUCAUCGUCAAGACCCACGACGAUGGAACCCCGGAGAAGCCCUUCGGACACGCCCUCGUC ........(((((((.....(((((.(((((.....))))))))))..((((....)))).((((((((.........))))))))....((((((.....))))))....))))).)). ( -47.80) >DroVir_CAF1 36980 120 - 1 AUUAUGAAACAGGGCAAAAUCGUAAUCGUCCUUAGUGGACGUUACGCCGGCCGUAAGGCCAUCAUUGUGAAGACCUCCGAUGAUGGUACGCCGGAGAAGCCCUUCGGUCAUGCGCUCGUU ..(((((...(((((.....(((((.(((((.....))))))))))((((((....)((((((((((.((.....))))))))))))..)))))....)))))....)))))........ ( -50.70) >DroGri_CAF1 34742 120 - 1 AUUAUGAAACAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGCCGCAAGGCCAUCAUCGUGAAGACCCAUGACGAUGGCACCCCAGAGAAGCCCUUCGGUCAUGCGCUCGUC ..(((((...(((((....((((((.(((((.....)))))))))(((((((....))))(((.(((((......))))).))))))......))...)))))....)))))........ ( -47.90) >DroWil_CAF1 33987 120 - 1 AUUAUGAAACAAGGCAAAAUCGUAAUCGUCCUUAGCGGACGUUACGCUGGCCGCAAAGCCAUCAUUGUCAAGACCUCUGAUGAUGGCACACCGGAGAAACCAUUCGGUCACGCUCUGGUU ............(((.....(((((.(((((.....))))))))))...))).....(((((((((............)))))))))..(((((((..(((....)))....))))))). ( -42.60) >DroMoj_CAF1 39948 120 - 1 AUUAUGAAACAGGGCAAGAUCGUAAUCGUCCUCAGUGGACGUUACGCCGGCCGCAAGGCUAUCAUUGUGAAGACUUCCGAUGAUGGCACGCCGGAGAAGCCCUUCGGUCAUGCACUCGUU ..(((((...(((((.....(((((.(((((.....))))))))))((((((....)((((((((((.((.....))))))))))))..)))))....)))))....)))))........ ( -51.20) >DroPer_CAF1 42454 120 - 1 AUCAUGAAGCAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCUGGCCGCAAGGCCAUCAUCGUCAAGACCUCCGACGAUGGCACCCCGGAGAAACCCUUCGGGCAUGCUCUGGUC .........(((((((.....((((.(((((.....)))))))))(((((((....))))).(((((((.........)))))))))..(((((((.....)))))))..)))))))... ( -54.80) >consensus AUUAUGAAACAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGCCGCAAGGCCAUCAUCGUCAAGACCUCCGACGAUGGCACCCCGGAGAAGCCCUUCGGUCAUGCGCUCGUC ...........((((.....(((((.(((((.....))))))))))..((((....))))...........).)))..(((((..((..(((((((.....)))))))...))..))))) (-35.74 = -35.47 + -0.27)



| Location | 21,510,135 – 21,510,242 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.33 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -25.09 |
| Energy contribution | -25.53 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21510135 107 - 27905053 -------------ACAGAGCAAACCGCCAAGAUGAGGAAAAUCAUGAAGCAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGUCGCAAGGCCAUCAUCGUCAAGACCCACGA -------------............((....((((......))))...)).((((..((((((((.(((((.....))))))))))..((((....))))......)))..).))).... ( -29.90) >DroVir_CAF1 37020 107 - 1 -------------GCAUGCACAGCCGCCAAGAUGAGGAAAAUUAUGAAACAGGGCAAAAUCGUAAUCGUCCUUAGUGGACGUUACGCCGGCCGUAAGGCCAUCAUUGUGAAGACCUCCGA -------------.....(((((.(((((...((((((..(((((((............)))))))..)))))).))).)).......((((....))))....)))))........... ( -32.10) >DroGri_CAF1 34782 101 - 1 -------------------ACAGCCGCCAAAAUGAGGAAAAUUAUGAAACAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGCCGCAAGGCCAUCAUCGUGAAGACCCAUGA -------------------...(((......((((......)))).......)))..((((((((.(((((.....))))))))))..((((....))))))).(((((......))))) ( -32.12) >DroWil_CAF1 34027 100 - 1 --------------------UAGCCUUCAAGAUGAGGAAAAUUAUGAAACAAGGCAAAAUCGUAAUCGUCCUUAGCGGACGUUACGCUGGCCGCAAAGCCAUCAUUGUCAAGACCUCUGA --------------------.............((((.......((...)).(((((...(((((.(((((.....)))))))))).((((......))))...)))))....))))... ( -24.30) >DroMoj_CAF1 39988 107 - 1 -------------ACACGCACAGCCGCCAAGAUGAGGAAAAUUAUGAAACAGGGCAAGAUCGUAAUCGUCCUCAGUGGACGUUACGCCGGCCGCAAGGCUAUCAUUGUGAAGACUUCCGA -------------.(((((..(((..(((...((((((..(((((((............)))))))..)))))).)))..)))..)).((((....))))......)))........... ( -33.20) >DroAna_CAF1 30883 120 - 1 UCAACAGCAGUAUCGAGACAAAGCCGCCAAAAUGAGGAAGAUUAUGAAACAGGGCAAGAUCGUCAUCGUCCUUAGCGGACGUUACGCCGGCCGCAAGGCCAUCAUCGUCAAGACCUCCGA ......((.((...........)).))......((((..(((.((((.....(((..(((....)))((((.....)))).....)))((((....)))).)))).)))....))))... ( -32.70) >consensus ______________CA__CACAGCCGCCAAGAUGAGGAAAAUUAUGAAACAGGGCAAGAUCGUAAUCGUCCUUAGCGGACGUUACGCCGGCCGCAAGGCCAUCAUCGUCAAGACCUCCGA .................................((((...((.((((.....(((......((((.(((((.....))))))))))))((((....)))).)))).)).....))))... (-25.09 = -25.53 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:30 2006