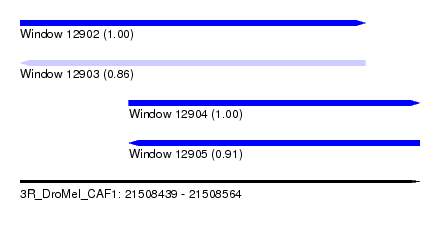
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,508,439 – 21,508,564 |
| Length | 125 |
| Max. P | 0.999082 |

| Location | 21,508,439 – 21,508,547 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 86.29 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -21.66 |
| Energy contribution | -21.26 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.87 |
| Structure conservation index | 0.79 |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21508439 108 + 27905053 CUUAAUCACUAACUUUUUGGUUGCCUUA----UA--UCUUUUAGUUUUAAGUACUUCUUCUUGGGUGACACAUGCUCGAUUGUCACCUAUAAGAGGCUACUUAAAAUUUGUUUA .........(((((....))))).....----..--......(((((((((((((((((..(((((((((..........))))))))).)))))).)))))))))))...... ( -28.60) >DroSec_CAF1 27996 108 + 1 CUUAAUCACUAACUUUUUGGUA----UAGUUUAA--UCUUUUAGUUUUAAGUACUUCUUCUUGGGUGACACAUGCUCGAUUGUCACCUACAAGAGGCCACUUAAAAUUUGUUUA .......(((((((....))).----))))....--......((((((((((.((((((..(((((((((..........))))))))).))))))..))))))))))...... ( -26.10) >DroSim_CAF1 29162 110 + 1 CUUAAUCACUAACUUUUUGGUUUCCUUAGUUU----CCUUUUAGUUUUAAGUACUUCUUCUUGGGUGACACAUGCUCGAUUGUCACCUACAAGAGGCCACUUAAAAUUUGUUUA .......(((((....)))))...........----......((((((((((.((((((..(((((((((..........))))))))).))))))..))))))))))...... ( -24.70) >DroEre_CAF1 28487 110 + 1 CUUAACCACUAACUAAUUGGUUUCCUUU----UAGUUGUUUUUGUUUUAAGUACUUCUUCAUGGGUGACACAUGCUCGUUAGUUACCUAUAAGAGGCCACUUAAAAUGUGUUUA ......((((((((((..((...))..)----)))))).....(((((((((.((((((.(((((((((((......))..)))))))))))))))..)))))))))))).... ( -31.20) >DroYak_CAF1 28834 108 + 1 CUUAACCACUAACUUUUUAGUUUCCUUA----UA--UCUUUUAGUUUUAAGUACUUCAUUAUGGGUAACACAUGCUCGUUGGUUACCUAUAAGAGGCCACUUAAAAUGUGUUUA ......(((.((((....))))......----..--.......(((((((((.((((.(((((((((((.((.......)))))))))))))))))..)))))))))))).... ( -27.10) >consensus CUUAAUCACUAACUUUUUGGUUUCCUUA____UA__UCUUUUAGUUUUAAGUACUUCUUCUUGGGUGACACAUGCUCGAUUGUCACCUAUAAGAGGCCACUUAAAAUUUGUUUA .......(((((....))))).....................((((((((((.((((((..((((((((............)))))))).))))))..))))))))))...... (-21.66 = -21.26 + -0.40)


| Location | 21,508,439 – 21,508,547 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 86.29 |
| Mean single sequence MFE | -22.16 |
| Consensus MFE | -15.82 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21508439 108 - 27905053 UAAACAAAUUUUAAGUAGCCUCUUAUAGGUGACAAUCGAGCAUGUGUCACCCAAGAAGAAGUACUUAAAACUAAAAGA--UA----UAAGGCAACCAAAAAGUUAGUGAUUAAG .........((((((((.(.((((...(((((((..........)))))))....)))).)))))))))(((((....--..----...(....).......)))))....... ( -21.29) >DroSec_CAF1 27996 108 - 1 UAAACAAAUUUUAAGUGGCCUCUUGUAGGUGACAAUCGAGCAUGUGUCACCCAAGAAGAAGUACUUAAAACUAAAAGA--UUAAACUA----UACCAAAAAGUUAGUGAUUAAG ........(((((((((.(.(((((..(((((((..........)))))))))))).)...)))))))))........--....((((----.((......))))))....... ( -20.90) >DroSim_CAF1 29162 110 - 1 UAAACAAAUUUUAAGUGGCCUCUUGUAGGUGACAAUCGAGCAUGUGUCACCCAAGAAGAAGUACUUAAAACUAAAAGG----AAACUAAGGAAACCAAAAAGUUAGUGAUUAAG .........((((((((.(.(((((..(((((((..........)))))))))))).)...))))))))(((((.((.----...))..(....).......)))))....... ( -25.30) >DroEre_CAF1 28487 110 - 1 UAAACACAUUUUAAGUGGCCUCUUAUAGGUAACUAACGAGCAUGUGUCACCCAUGAAGAAGUACUUAAAACAAAAACAACUA----AAAGGAAACCAAUUAGUUAGUGGUUAAG ....(((.(((((((((.(.((((...(((.((............)).)))....)))).)))))))))).......(((((----(..(....)...)))))).)))...... ( -20.60) >DroYak_CAF1 28834 108 - 1 UAAACACAUUUUAAGUGGCCUCUUAUAGGUAACCAACGAGCAUGUGUUACCCAUAAUGAAGUACUUAAAACUAAAAGA--UA----UAAGGAAACUAAAAAGUUAGUGGUUAAG ....(((.(((((((((.(.((((((.((((((((.......)).)))))).)))).)).))))))))))........--..----..((....)).........)))...... ( -22.70) >consensus UAAACAAAUUUUAAGUGGCCUCUUAUAGGUGACAAUCGAGCAUGUGUCACCCAAGAAGAAGUACUUAAAACUAAAAGA__UA____UAAGGAAACCAAAAAGUUAGUGAUUAAG ..(((...(((((((((.(.((((...((((((............))))))....)))).))))))))))...................(....)......))).......... (-15.82 = -16.02 + 0.20)

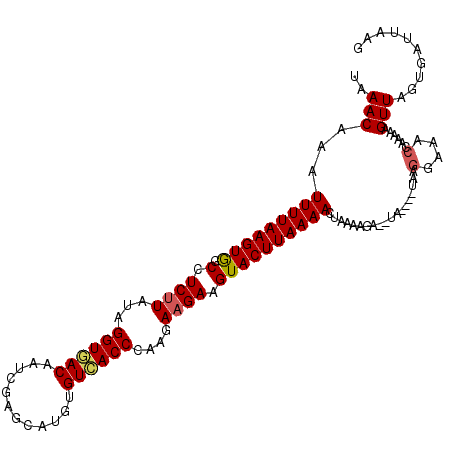
| Location | 21,508,473 – 21,508,564 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 90.20 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -20.64 |
| Energy contribution | -20.40 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.999082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21508473 91 + 27905053 UUAGUUUUAAGUACUUCUUCUUGGGUGACACAUGCUCGAUUGUCACCUAUAAGAGGCUACUUAAAAUUUGUUUAUAUAAA--UUGGACUUAUG ..(((((((((((((((((..(((((((((..........))))))))).)))))).))))))))))).(((((......--.)))))..... ( -28.50) >DroSec_CAF1 28030 91 + 1 UUAGUUUUAAGUACUUCUUCUUGGGUGACACAUGCUCGAUUGUCACCUACAAGAGGCCACUUAAAAUUUGUUUAUAUAAA--UUGGACCACUG ..((((((((((.((((((..(((((((((..........))))))))).))))))..)))))))))).(((((......--.)))))..... ( -25.00) >DroSim_CAF1 29198 91 + 1 UUAGUUUUAAGUACUUCUUCUUGGGUGACACAUGCUCGAUUGUCACCUACAAGAGGCCACUUAAAAUUUGUUUAUAUAAA--UUGGACCACUG ..((((((((((.((((((..(((((((((..........))))))))).))))))..)))))))))).(((((......--.)))))..... ( -25.00) >DroEre_CAF1 28523 91 + 1 UUUGUUUUAAGUACUUCUUCAUGGGUGACACAUGCUCGUUAGUUACCUAUAAGAGGCCACUUAAAAUGUGUUUAUAUAAA--UUCGACCACUG ...(((((((((.((((((.(((((((((((......))..)))))))))))))))..))))))))).............--........... ( -23.30) >DroYak_CAF1 28868 93 + 1 UUAGUUUUAAGUACUUCAUUAUGGGUAACACAUGCUCGUUGGUUACCUAUAAGAGGCCACUUAAAAUGUGUUUAUAUAAUUUUUCGACAACUG ...(((((((((.((((.(((((((((((.((.......)))))))))))))))))..))))))))).......................... ( -25.10) >consensus UUAGUUUUAAGUACUUCUUCUUGGGUGACACAUGCUCGAUUGUCACCUAUAAGAGGCCACUUAAAAUUUGUUUAUAUAAA__UUGGACCACUG ..((((((((((.((((((..((((((((............)))))))).))))))..))))))))))......................... (-20.64 = -20.40 + -0.24)


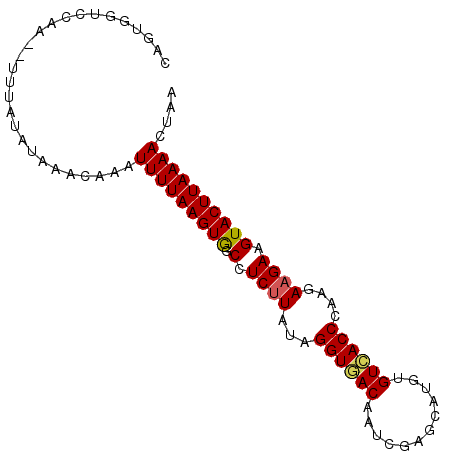
| Location | 21,508,473 – 21,508,564 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 90.20 |
| Mean single sequence MFE | -18.02 |
| Consensus MFE | -15.72 |
| Energy contribution | -15.72 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21508473 91 - 27905053 CAUAAGUCCAA--UUUAUAUAAACAAAUUUUAAGUAGCCUCUUAUAGGUGACAAUCGAGCAUGUGUCACCCAAGAAGAAGUACUUAAAACUAA ...........--..............(((((((((.(.((((...(((((((..........)))))))....)))).)))))))))).... ( -19.30) >DroSec_CAF1 28030 91 - 1 CAGUGGUCCAA--UUUAUAUAAACAAAUUUUAAGUGGCCUCUUGUAGGUGACAAUCGAGCAUGUGUCACCCAAGAAGAAGUACUUAAAACUAA ...........--..............(((((((((.(.(((((..(((((((..........)))))))))))).)...))))))))).... ( -19.90) >DroSim_CAF1 29198 91 - 1 CAGUGGUCCAA--UUUAUAUAAACAAAUUUUAAGUGGCCUCUUGUAGGUGACAAUCGAGCAUGUGUCACCCAAGAAGAAGUACUUAAAACUAA ...........--..............(((((((((.(.(((((..(((((((..........)))))))))))).)...))))))))).... ( -19.90) >DroEre_CAF1 28523 91 - 1 CAGUGGUCGAA--UUUAUAUAAACACAUUUUAAGUGGCCUCUUAUAGGUAACUAACGAGCAUGUGUCACCCAUGAAGAAGUACUUAAAACAAA ..(((......--..........))).(((((((((.(.((((...(((.((............)).)))....)))).)))))))))).... ( -14.09) >DroYak_CAF1 28868 93 - 1 CAGUUGUCGAAAAAUUAUAUAAACACAUUUUAAGUGGCCUCUUAUAGGUAACCAACGAGCAUGUGUUACCCAUAAUGAAGUACUUAAAACUAA ...........................(((((((((.(.((((((.((((((((.......)).)))))).)))).)).)))))))))).... ( -16.90) >consensus CAGUGGUCCAA__UUUAUAUAAACAAAUUUUAAGUGGCCUCUUAUAGGUGACAAUCGAGCAUGUGUCACCCAAGAAGAAGUACUUAAAACUAA ...........................(((((((((.(.((((...((((((............))))))....)))).)))))))))).... (-15.72 = -15.72 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:28 2006