
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,467,964 – 21,468,075 |
| Length | 111 |
| Max. P | 0.995486 |

| Location | 21,467,964 – 21,468,075 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -21.54 |
| Consensus MFE | -5.00 |
| Energy contribution | -6.72 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.23 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.731114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21467964 111 + 27905053 AUUUAUGUAU---------AAUAGUUGAAUGCGGAAUUGCAAGUACGCCUUCAACUUUUAACUGCCAUGUUUCGGUUAAAUCCCAAUUCUCUAAGGACAGUUCCCUAGAGGAAAUUGCAU ....(((((.---------...(((((((.(((............))).)))))))((((((((........))))))))......(((((((.(((....))).)))))))...))))) ( -27.90) >DroSec_CAF1 74727 108 + 1 ---AAUGUAU---------AUUAGUUGAAUGCGGAAUUGCAAGUUCCCAUUCAACUUUUAAUUGCCAUCUUCGAAUUAAUUCCCAAUUCCCUUAGGACAAUUCCCCAGAGAAAACUUUAU ---......(---------(..(((((((((.(((((.....))))))))))))))..))..........................(((.((..(((....)))..)).)))........ ( -21.50) >DroSim_CAF1 59631 108 + 1 ---AAUGUAU---------AUUAGUUGAAUGCGGAAUUCCAAGUUCCCAUUCAACUUUUAAUUGCCAUCUUCCGAUUAAUUCCCAAUUCCCUUAGGACAAUUCCCCAGAGAAAACUGUAU ---.......---------...(((((((((.(((((.....)))))))))))))).(((((((........))))))).......(((.((..(((....)))..)).)))........ ( -22.50) >DroEre_CAF1 75899 99 + 1 ---AAUGUAUGUAUAUACUAAAAGUUGAAUACGGAAUUGCAAAUU------------------GCCAUGUUCCGGUCAAUUCCCAAUUCGCAAAGGACAAUUCCCCAGUGGAAAUUGUAU ---...........((((....((((((...((((((.((.....------------------))...)))))).)))))).....(((((...(((....)))...)))))....)))) ( -19.90) >DroYak_CAF1 66542 106 + 1 ---AAUGUAU---------ACAAGUUGAAUGCGAAAUUGCCAAUUCACAUGCAACUUUUUAAUGCCAUG-UCCGGUUAAUUCCCAAUUCUCCAAAGACAAUUCCCCAGUGGAA-CUGUAU ---...((((---------..((((((.(((.(((........))).))).))))))....))))....-...((..(((.....)))..))....(((.((((.....))))-.))).. ( -15.90) >consensus ___AAUGUAU_________AAUAGUUGAAUGCGGAAUUGCAAGUUCCCAUUCAACUUUUAAUUGCCAUGUUCCGGUUAAUUCCCAAUUCCCUAAGGACAAUUCCCCAGAGGAAACUGUAU ......................((((...((.(((((((..................(((((((........)))))))........(((....)))))))))).)).....)))).... ( -5.00 = -6.72 + 1.72)



| Location | 21,467,964 – 21,468,075 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -11.10 |
| Energy contribution | -11.69 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.39 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21467964 111 - 27905053 AUGCAAUUUCCUCUAGGGAACUGUCCUUAGAGAAUUGGGAUUUAACCGAAACAUGGCAGUUAAAAGUUGAAGGCGUACUUGCAAUUCCGCAUUCAACUAUU---------AUACAUAAAU .(((......(((((((((....)))))))))..((((.......))))......)))(((((.(((((((.(((............))).))))))).))---------).))...... ( -30.00) >DroSec_CAF1 74727 108 - 1 AUAAAGUUUUCUCUGGGGAAUUGUCCUAAGGGAAUUGGGAAUUAAUUCGAAGAUGGCAAUUAAAAGUUGAAUGGGAACUUGCAAUUCCGCAUUCAACUAAU---------AUACAUU--- .....((..(((.((((......((((((.....)))))).....)))).)))..)).......(((((((((((((.......)))).)))))))))...---------.......--- ( -27.40) >DroSim_CAF1 59631 108 - 1 AUACAGUUUUCUCUGGGGAAUUGUCCUAAGGGAAUUGGGAAUUAAUCGGAAGAUGGCAAUUAAAAGUUGAAUGGGAACUUGGAAUUCCGCAUUCAACUAAU---------AUACAUU--- ...(((((..((.(((((.....)))))))..))))).......(((....)))..........(((((((((((((.......)))).)))))))))...---------.......--- ( -30.30) >DroEre_CAF1 75899 99 - 1 AUACAAUUUCCACUGGGGAAUUGUCCUUUGCGAAUUGGGAAUUGACCGGAACAUGGC------------------AAUUUGCAAUUCCGUAUUCAACUUUUAGUAUAUACAUACAUU--- ..(((((((((....)))))))))....(((((((((.((((((.(((.....))))------------------))))).))))).))))...........((((....))))...--- ( -26.80) >DroYak_CAF1 66542 106 - 1 AUACAG-UUCCACUGGGGAAUUGUCUUUGGAGAAUUGGGAAUUAACCGGA-CAUGGCAUUAAAAAGUUGCAUGUGAAUUGGCAAUUUCGCAUUCAACUUGU---------AUACAUU--- ((((((-(.(((((((..((((.((..(.....)..)).))))..)))).-..))).)))...((((((.(((((((........))))))).))))))))---------)).....--- ( -26.70) >consensus AUACAGUUUCCUCUGGGGAAUUGUCCUUAGAGAAUUGGGAAUUAACCGGAACAUGGCAAUUAAAAGUUGAAUGGGAACUUGCAAUUCCGCAUUCAACUAAU_________AUACAUU___ ..(((((((((....))))))))).......((((..((((((.............((((.....)))).............))))))..)))).......................... (-11.10 = -11.69 + 0.59)

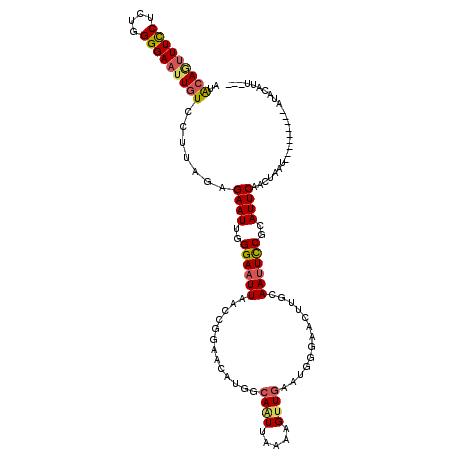

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:00:01 2006