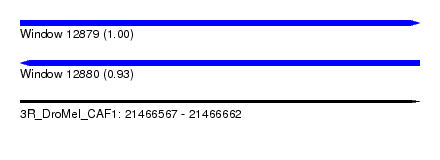
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,466,567 – 21,466,662 |
| Length | 95 |
| Max. P | 0.998458 |

| Location | 21,466,567 – 21,466,662 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 90.76 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -24.95 |
| Energy contribution | -24.98 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.998458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
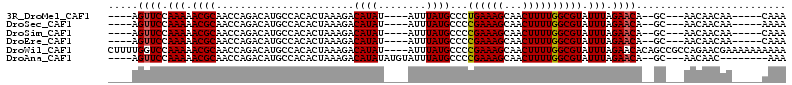
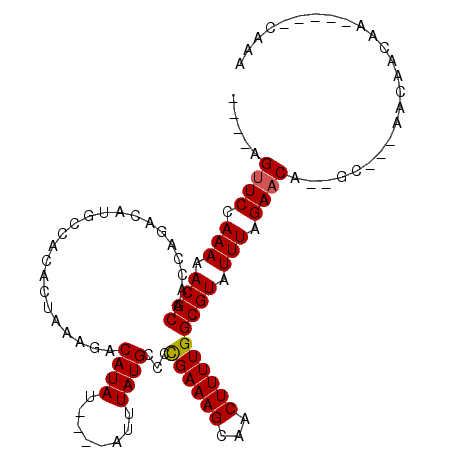
>3R_DroMel_CAF1 21466567 95 + 27905053 UUUG-----UUGUUGUU---GC--UGUUCUAAAUACGCCAAAAGUUGCUUUCAGGGCAUAAAU----AUAUGUCUUUAGUGUGGCAUGUCUGGUUGCGUUUUUGGAACU---- ....-----........---..--.((((((((.((((.(..((.((((...((((((((...----.))))))))......))))...))..).)))).)))))))).---- ( -25.50) >DroSec_CAF1 73349 95 + 1 UUUU-----UUGUUGUU---GC--UGUUCUAAAUACGCCAAAAGUUGCUUUCGGGGCAUAAAU----AUAUGUCUUUAGUGUGGCAUGUCUGGUUGCGUUUUUGGAACU---- ....-----........---..--.((((((((.((((.(..((.((((..(.(((((((...----.)))))))...)...))))...))..).)))).)))))))).---- ( -25.60) >DroSim_CAF1 58253 95 + 1 UUUG-----UUGUUGUU---GC--UGUUCUAAAUACGCCAAAAGUUGCUUUCGGGGCAUAAAU----AUAUGUCUUUAGUGUGGCAUGUCUGGUUGCGUUUUUGGAACU---- ....-----........---..--.((((((((.((((.(..((.((((..(.(((((((...----.)))))))...)...))))...))..).)))).)))))))).---- ( -25.60) >DroEre_CAF1 74508 95 + 1 UUUG-----UUGUUGUU---GC--UGUUCUAAAUACGCCAAAAGUUGCUUUCGGGGCAUAAAU----AUAUGUCUUUAGUGUGGCAUGUCUGGUUGCGUUUUUGGAACU---- ....-----........---..--.((((((((.((((.(..((.((((..(.(((((((...----.)))))))...)...))))...))..).)))).)))))))).---- ( -25.60) >DroWil_CAF1 170426 109 + 1 UUUUUUUUUUCGUUCUGGCGGCUGUGUUCUAAAUACGCCAAAAGUUGCUUUCGGGGCAUAAAU----AUAUGUCUUUAGUGUGGCAUGUCUGGUUGCGUUUUUGGACCAAAAG ................((((((.((((.....)))))))....((..(.((..(((((((...----.)))))))..)).)..))..)))(((((.((....))))))).... ( -25.20) >DroAna_CAF1 153476 96 + 1 UUU--------GUUGUU---GC--UGUUCUAAAUACGCCAAAAGUUGCUUUCGGGGCAUAAAUACAUAUAUGUCUUUAGUGUGGCAUGUCUGGUUGCGUUUUUGGAACU---- ...--------......---..--.((((((((.((((.(..((.((((..(.(((((((........)))))))...)...))))...))..).)))).)))))))).---- ( -25.60) >consensus UUUG_____UUGUUGUU___GC__UGUUCUAAAUACGCCAAAAGUUGCUUUCGGGGCAUAAAU____AUAUGUCUUUAGUGUGGCAUGUCUGGUUGCGUUUUUGGAACU____ .........................((((((((.((((.(..((.((((...((((((((........))))))))......))))...))..).)))).))))))))..... (-24.95 = -24.98 + 0.03)


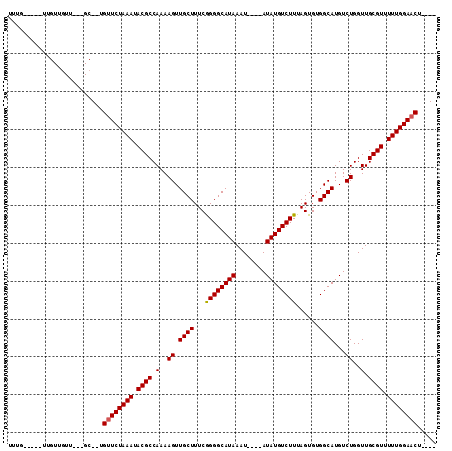
| Location | 21,466,567 – 21,466,662 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 90.76 |
| Mean single sequence MFE | -14.99 |
| Consensus MFE | -13.84 |
| Energy contribution | -13.87 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21466567 95 - 27905053 ----AGUUCCAAAAACGCAACCAGACAUGCCACACUAAAGACAUAU----AUUUAUGCCCUGAAAGCAACUUUUGGCGUAUUUAGAACA--GC---AACAACAA-----CAAA ----.((((.......(((........)))...............(----(..((((((..(((((...)))))))))))..)))))).--..---........-----.... ( -13.40) >DroSec_CAF1 73349 95 - 1 ----AGUUCCAAAAACGCAACCAGACAUGCCACACUAAAGACAUAU----AUUUAUGCCCCGAAAGCAACUUUUGGCGUAUUUAGAACA--GC---AACAACAA-----AAAA ----.((((.(((.((((.......................((((.----...))))...((((((...)))))))))).))).)))).--..---........-----.... ( -14.90) >DroSim_CAF1 58253 95 - 1 ----AGUUCCAAAAACGCAACCAGACAUGCCACACUAAAGACAUAU----AUUUAUGCCCCGAAAGCAACUUUUGGCGUAUUUAGAACA--GC---AACAACAA-----CAAA ----.((((.(((.((((.......................((((.----...))))...((((((...)))))))))).))).)))).--..---........-----.... ( -14.90) >DroEre_CAF1 74508 95 - 1 ----AGUUCCAAAAACGCAACCAGACAUGCCACACUAAAGACAUAU----AUUUAUGCCCCGAAAGCAACUUUUGGCGUAUUUAGAACA--GC---AACAACAA-----CAAA ----.((((.(((.((((.......................((((.----...))))...((((((...)))))))))).))).)))).--..---........-----.... ( -14.90) >DroWil_CAF1 170426 109 - 1 CUUUUGGUCCAAAAACGCAACCAGACAUGCCACACUAAAGACAUAU----AUUUAUGCCCCGAAAGCAACUUUUGGCGUAUUUAGAACACAGCCGCCAGAACGAAAAAAAAAA (((((((.........(((........)))...........((((.----...))))..)))))))....((((((((...............))))))))............ ( -16.96) >DroAna_CAF1 153476 96 - 1 ----AGUUCCAAAAACGCAACCAGACAUGCCACACUAAAGACAUAUAUGUAUUUAUGCCCCGAAAGCAACUUUUGGCGUAUUUAGAACA--GC---AACAAC--------AAA ----.((((.(((.((((.......................((((........))))...((((((...)))))))))).))).)))).--..---......--------... ( -14.90) >consensus ____AGUUCCAAAAACGCAACCAGACAUGCCACACUAAAGACAUAU____AUUUAUGCCCCGAAAGCAACUUUUGGCGUAUUUAGAACA__GC___AACAACAA_____CAAA .....((((.(((.((((.......................((((........))))...((((((...)))))))))).))).))))......................... (-13.84 = -13.87 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:59 2006