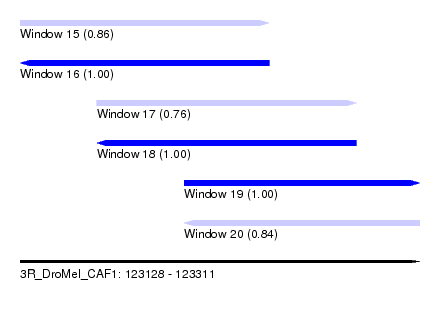
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 123,128 – 123,311 |
| Length | 183 |
| Max. P | 0.999334 |

| Location | 123,128 – 123,242 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.44 |
| Mean single sequence MFE | -16.95 |
| Consensus MFE | -16.10 |
| Energy contribution | -16.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.857869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 123128 114 + 27905053 AGUU-AACAUG--UC--AAACAUUUGUAGUCUACUCUACAAAAUAUGCUUAUUUAUUGCUGUUUCCAUACUCAAUUUCUU-UUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUG ....-..((((--(.--.....(((((((......))))))))))))..((((((((((.(((((..........((((.-....)))).(((....)))....))))))).)))))))) ( -16.40) >DroVir_CAF1 34257 116 + 1 AGUUGAACAUA--CA--UAACAUUUGUAGUCUACUCUACAAAAUAUGCUUAUUUAUUGCUGUUUCCAUACUCAAUUUCUUUUUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUG ...........--((--((...(((((((......))))))).))))..((((((((((.(((((..........((((......)))).(((....)))....))))))).)))))))) ( -16.80) >DroGri_CAF1 34089 116 + 1 AGUUGAACAUA--CA--UAACAUUUGUAGUCUACUCUACAAAAUAUGCUUAUUUAUUGCUGUUUCCAUACUCAAUUUCUUUUUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUG ...........--((--((...(((((((......))))))).))))..((((((((((.(((((..........((((......)))).(((....)))....))))))).)))))))) ( -16.80) >DroWil_CAF1 48963 117 + 1 AGUU-GACAUACAUA--AAACAUUUGUAGUCUACUCUACAAAAUAUGCUUAUUUAUUGCUGUUUCCAUACUCAAUUUCUUUUUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUG ....-..........--.....(((((((......))))))).......((((((((((.(((((..........((((......)))).(((....)))....))))))).)))))))) ( -16.10) >DroMoj_CAF1 29779 118 + 1 AGUUAAACAUA--CAUAUAACAUUUGUAGUCUACUCUACAAAGUAUGCUUAUUUAUUGCUGUUUCCAUACUCAAUUUCUUUUUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUG .......((((--(........(((((((......))))))))))))..((((((((((.(((((..........((((......)))).(((....)))....))))))).)))))))) ( -17.90) >DroAna_CAF1 22515 114 + 1 AGUU-AACAUA--UU--UAACAUUUGUAGUCUACUCUACAAAAUAUGCUUAUUUAUUGCUGUUUCCAUACUCAAUUUCUU-UUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUG .(((-((....--.)--)))).(((((((......))))))).......((((((((((.(((((..........((((.-....)))).(((....)))....))))))).)))))))) ( -17.70) >consensus AGUU_AACAUA__CA__UAACAUUUGUAGUCUACUCUACAAAAUAUGCUUAUUUAUUGCUGUUUCCAUACUCAAUUUCUUUUUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUG ......................(((((((......))))))).......((((((((((.(((((..........((((......)))).(((....)))....))))))).)))))))) (-16.10 = -16.10 + -0.00)

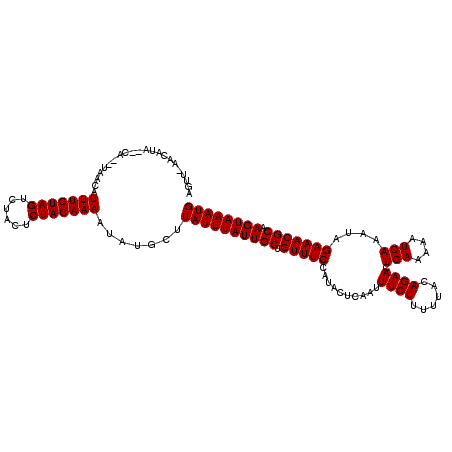
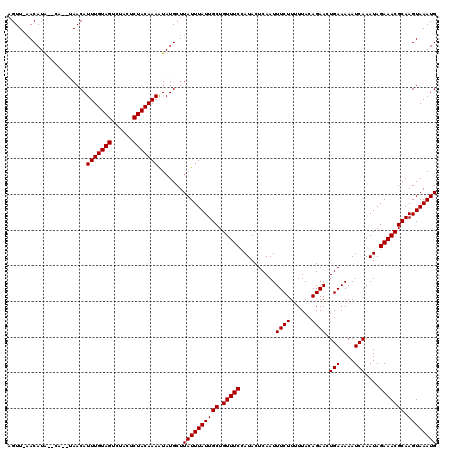
| Location | 123,128 – 123,242 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.44 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -23.70 |
| Energy contribution | -23.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 123128 114 - 27905053 CAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAA-AAGAAAUUGAGUAUGGAAACAGCAAUAAAUAAGCAUAUUUUGUAGAGUAGACUACAAAUGUUU--GA--CAUGUU-AACU .(((((.((((((((((((.......(((((((((....-.)).))))))).)))))))).)))))))))((((((...((((((......))))))))))))--..--......-.... ( -24.50) >DroVir_CAF1 34257 116 - 1 CAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAAAAAGAAAUUGAGUAUGGAAACAGCAAUAAAUAAGCAUAUUUUGUAGAGUAGACUACAAAUGUUA--UG--UAUGUUCAACU .(((((.((((((((((((.......(((((((((......)).))))))).)))))))).)))))))))..(((((.(((((((......)))))))...))--))--).......... ( -27.00) >DroGri_CAF1 34089 116 - 1 CAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAAAAAGAAAUUGAGUAUGGAAACAGCAAUAAAUAAGCAUAUUUUGUAGAGUAGACUACAAAUGUUA--UG--UAUGUUCAACU .(((((.((((((((((((.......(((((((((......)).))))))).)))))))).)))))))))..(((((.(((((((......)))))))...))--))--).......... ( -27.00) >DroWil_CAF1 48963 117 - 1 CAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAAAAAGAAAUUGAGUAUGGAAACAGCAAUAAAUAAGCAUAUUUUGUAGAGUAGACUACAAAUGUUU--UAUGUAUGUC-AACU .(((((.((((((((((((.......(((((((((......)).))))))).)))))))).)))))))))..(((((.(((((((......))))))).....--))))).....-.... ( -25.70) >DroMoj_CAF1 29779 118 - 1 CAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAAAAAGAAAUUGAGUAUGGAAACAGCAAUAAAUAAGCAUACUUUGUAGAGUAGACUACAAAUGUUAUAUG--UAUGUUUAACU ..((((.((((((((((((.......(((((((((......)).))))))).)))))))).))))))))((((((((((((((((......)))))))........)--))))))))... ( -28.40) >DroAna_CAF1 22515 114 - 1 CAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAA-AAGAAAUUGAGUAUGGAAACAGCAAUAAAUAAGCAUAUUUUGUAGAGUAGACUACAAAUGUUA--AA--UAUGUU-AACU .(((((.((((((((((((.......(((((((((....-.)).))))))).)))))))).))))))))).((((((((((((((......)))))......)--))--))))))-.... ( -25.80) >consensus CAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAAAAAGAAAUUGAGUAUGGAAACAGCAAUAAAUAAGCAUAUUUUGUAGAGUAGACUACAAAUGUUA__UA__UAUGUU_AACU .(((((.((((((((((((.......(((((((((......)).))))))).)))))))).))))))))).(((((...((((((......))))))))))).................. (-23.70 = -23.70 + -0.00)

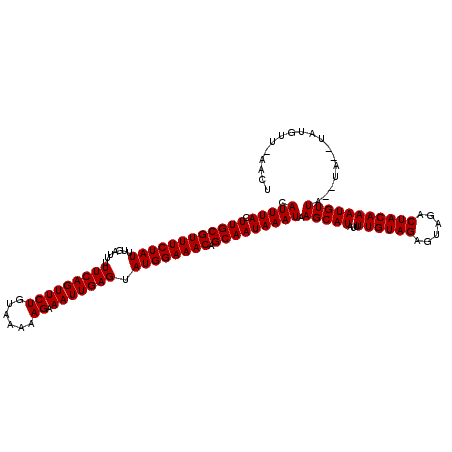
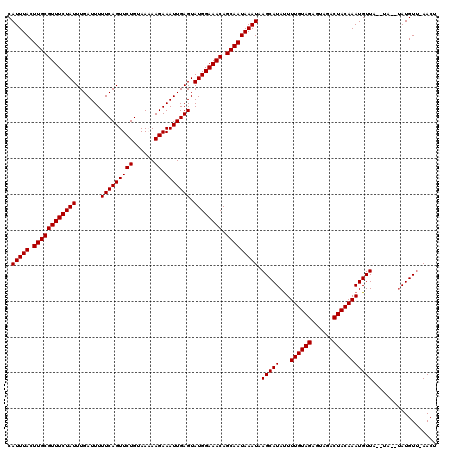
| Location | 123,163 – 123,282 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -19.94 |
| Energy contribution | -19.80 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 123163 119 + 27905053 AAAUAUGCUUAUUUAUUGCUGUUUCCAUACUCAAUUUCUU-UUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUGAGCUAAAAACUCAAAUAGAGGGAAAAUGUGUCAUGCAGUG ......(((((((((((((.(((((..........((((.-....)))).(((....)))....))))))).)))))))))))......(((.....)))......(((.....)))... ( -20.50) >DroVir_CAF1 34293 120 + 1 AAAUAUGCUUAUUUAUUGCUGUUUCCAUACUCAAUUUCUUUUUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUGAGCUAAAAACUCAAAUAGAGGGAAAAUGUGUCAUGCAGUG ......(((((((((((((.(((((..........((((......)))).(((....)))....))))))).)))))))))))......(((.....)))......(((.....)))... ( -20.70) >DroGri_CAF1 34125 120 + 1 AAAUAUGCUUAUUUAUUGCUGUUUCCAUACUCAAUUUCUUUUUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUGAGCUAAAAACUCAAAUAGAGGGAAAAUGUGUCAUGCAGUG ......(((((((((((((.(((((..........((((......)))).(((....)))....))))))).)))))))))))......(((.....)))......(((.....)))... ( -20.70) >DroWil_CAF1 49000 120 + 1 AAAUAUGCUUAUUUAUUGCUGUUUCCAUACUCAAUUUCUUUUUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUGAGCUAAAAACCCAAAUAGAGGGAAAAUGUGUCAUGCAGUG ......(((((((((((((.(((((..........((((......)))).(((....)))....))))))).)))))))))))......(((.......)))....(((.....)))... ( -22.30) >DroYak_CAF1 29885 119 + 1 AAAUAUGCUUAUUUAUUGCUGUUUCCAUACUCAAUUUCUU-UUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUGAGCUAAAAACUCAAAUAGAGGGAAAAUGUGUCAUGCAGUG ......(((((((((((((.(((((..........((((.-....)))).(((....)))....))))))).)))))))))))......(((.....)))......(((.....)))... ( -20.50) >DroMoj_CAF1 29817 120 + 1 AAGUAUGCUUAUUUAUUGCUGUUUCCAUACUCAAUUUCUUUUUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUGAGCUAAAAACUCAAAUAGAGGGAAAAUGUGUCAUGCAGUG ..(((((((((((((((((.(((((..........((((......)))).(((....)))....))))))).)))))))))))......(((.....)))............)))).... ( -22.80) >consensus AAAUAUGCUUAUUUAUUGCUGUUUCCAUACUCAAUUUCUUUUUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUGAGCUAAAAACUCAAAUAGAGGGAAAAUGUGUCAUGCAGUG ......(((((((((((((.(((((..........((((......)))).(((....)))....))))))).)))))))))))......(((.......)))....(((.....)))... (-19.94 = -19.80 + -0.14)

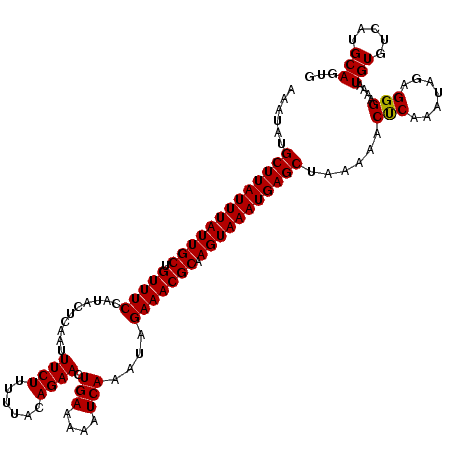
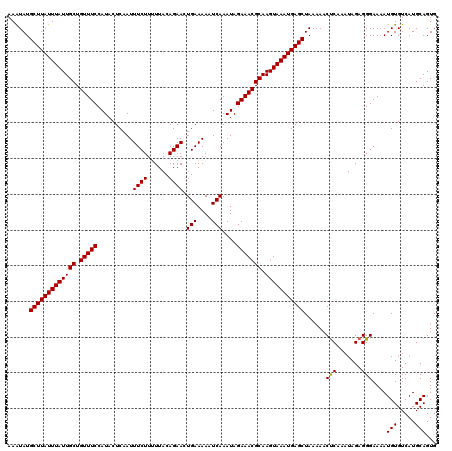
| Location | 123,163 – 123,282 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -23.99 |
| Energy contribution | -23.85 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.999192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 123163 119 - 27905053 CACUGCAUGACACAUUUUCCCUCUAUUUGAGUUUUUAGCUCAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAA-AAGAAAUUGAGUAUGGAAACAGCAAUAAAUAAGCAUAUUU ....................(((.....)))......(((.(((((.((((((((((((.......(((((((((....-.)).))))))).)))))))).))))))))).)))...... ( -23.80) >DroVir_CAF1 34293 120 - 1 CACUGCAUGACACAUUUUCCCUCUAUUUGAGUUUUUAGCUCAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAAAAAGAAAUUGAGUAUGGAAACAGCAAUAAAUAAGCAUAUUU ....................(((.....)))......(((.(((((.((((((((((((.......(((((((((......)).))))))).)))))))).))))))))).)))...... ( -24.00) >DroGri_CAF1 34125 120 - 1 CACUGCAUGACACAUUUUCCCUCUAUUUGAGUUUUUAGCUCAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAAAAAGAAAUUGAGUAUGGAAACAGCAAUAAAUAAGCAUAUUU ....................(((.....)))......(((.(((((.((((((((((((.......(((((((((......)).))))))).)))))))).))))))))).)))...... ( -24.00) >DroWil_CAF1 49000 120 - 1 CACUGCAUGACACAUUUUCCCUCUAUUUGGGUUUUUAGCUCAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAAAAAGAAAUUGAGUAUGGAAACAGCAAUAAAUAAGCAUAUUU ..................(((.......)))......(((.(((((.((((((((((((.......(((((((((......)).))))))).)))))))).))))))))).)))...... ( -25.60) >DroYak_CAF1 29885 119 - 1 CACUGCAUGACACAUUUUCCCUCUAUUUGAGUUUUUAGCUCAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAA-AAGAAAUUGAGUAUGGAAACAGCAAUAAAUAAGCAUAUUU ....................(((.....)))......(((.(((((.((((((((((((.......(((((((((....-.)).))))))).)))))))).))))))))).)))...... ( -23.80) >DroMoj_CAF1 29817 120 - 1 CACUGCAUGACACAUUUUCCCUCUAUUUGAGUUUUUAGCUCAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAAAAAGAAAUUGAGUAUGGAAACAGCAAUAAAUAAGCAUACUU ....................(((.....)))......(((.(((((.((((((((((((.......(((((((((......)).))))))).)))))))).))))))))).)))...... ( -24.00) >consensus CACUGCAUGACACAUUUUCCCUCUAUUUGAGUUUUUAGCUCAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAAAAAGAAAUUGAGUAUGGAAACAGCAAUAAAUAAGCAUAUUU ....................(((.....)))......(((.(((((.((((((((((((.......(((((((((......)).))))))).)))))))).))))))))).)))...... (-23.99 = -23.85 + -0.14)

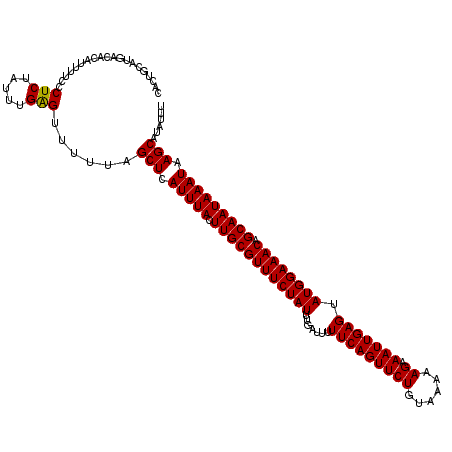
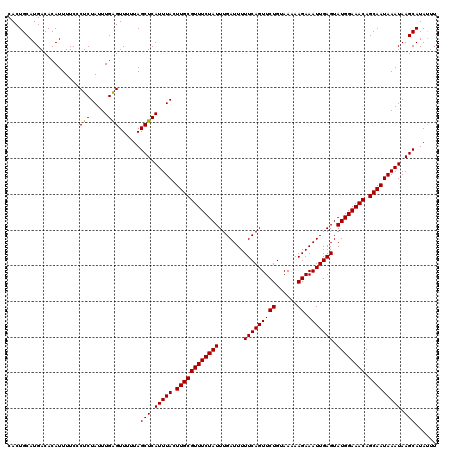
| Location | 123,203 – 123,311 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 94.02 |
| Mean single sequence MFE | -16.32 |
| Consensus MFE | -16.04 |
| Energy contribution | -16.04 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 123203 108 + 27905053 -UUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUGAGCUAAAAACUCAAAUAGAGGGAAAAUGUGUCAUGCAGUGAUAUGUCAGUCACCAUAAUAACAG-AUAAA -........(((.....((........((....))...((((.......))))....))((((..(((((((((...)))))))))...)).)).......)))-..... ( -16.90) >DroVir_CAF1 34333 109 + 1 UUUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUGAGCUAAAAACUCAAAUAGAGGGAAAAUGUGUCAUGCAGUGAUAUGUCAGUCAUCAUAAAUGAUA-UAUAA ......((.((((...((((.......((((((((......))).....(((.....)))......))))).......))))...))))))((((....)))).-..... ( -16.84) >DroPse_CAF1 27263 109 + 1 -UUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUGAGCUAAAAACUCAAAUAGAGGGAAAAUGUGUCAUGCAGUGAUAUGUCAGUCAUCAUAAAAGCAAAAUAAA -.....((.((((...((((.......((((((((......))).....(((.....)))......))))).......))))...))))))................... ( -16.04) >DroGri_CAF1 34165 109 + 1 UUUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUGAGCUAAAAACUCAAAUAGAGGGAAAAUGUGUCAUGCAGUGAUAUGUCAGUCAUCAUAAAAGAAA-AAAAA ......((.((((...((((.......((((((((......))).....(((.....)))......))))).......))))...)))))).............-..... ( -16.04) >DroAna_CAF1 22590 108 + 1 -UUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUGAGCUAAAAACUCAAAUAGAGGGAAAAUGUGUCAUGCAGUGAUAUGUCAGUCAACAAAAUAAAAG-CGAAA -.....((.((((...((((.......((((((((......))).....(((.....)))......))))).......))))...)))))).............-..... ( -16.04) >DroPer_CAF1 26284 109 + 1 -UUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUGAGCUAAAAACUCAAAUAGAGGGAAAAUGUGUCAUGCAGUGAUAUGUCAGUCAUCAUAAAAGCAAAAUAAA -.....((.((((...((((.......((((((((......))).....(((.....)))......))))).......))))...))))))................... ( -16.04) >consensus _UUACAGAACUGAAAAAUCAAAUAGAAACGCAAGUAAAUGAGCUAAAAACUCAAAUAGAGGGAAAAUGUGUCAUGCAGUGAUAUGUCAGUCAUCAUAAAAGAAA_AUAAA ......((.((((...((((.......((((((((......))).....(((.....)))......))))).......))))...))))))................... (-16.04 = -16.04 + -0.00)

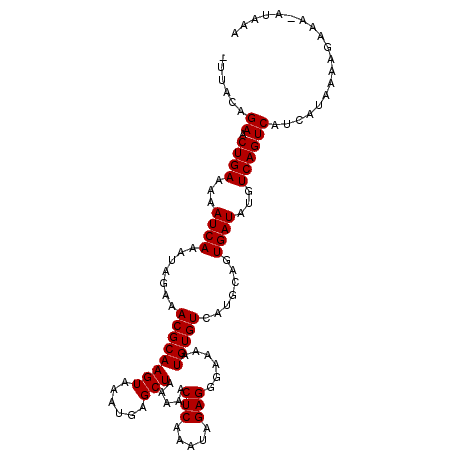
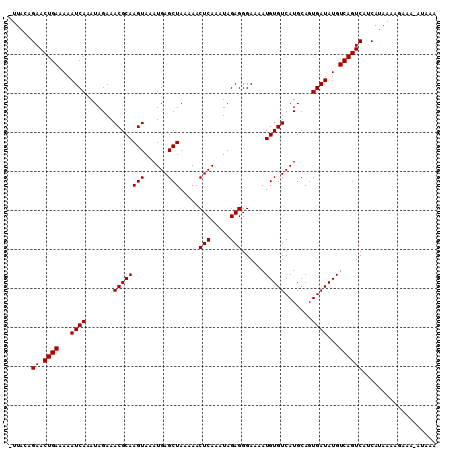
| Location | 123,203 – 123,311 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 94.02 |
| Mean single sequence MFE | -14.75 |
| Consensus MFE | -13.90 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 123203 108 - 27905053 UUUAU-CUGUUAUUAUGGUGACUGACAUAUCACUGCAUGACACAUUUUCCCUCUAUUUGAGUUUUUAGCUCAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAA- .....-......((((((.((((((...((((......(((.((.........((..(((((.....)))))..))..)).)))......))))...))))))))))))- ( -17.00) >DroVir_CAF1 34333 109 - 1 UUAUA-UAUCAUUUAUGAUGACUGACAUAUCACUGCAUGACACAUUUUCCCUCUAUUUGAGUUUUUAGCUCAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAAA (((((-(((((....)))))(((((...((((......(((.((.........((..(((((.....)))))..))..)).)))......))))...)))))..))))). ( -15.20) >DroPse_CAF1 27263 109 - 1 UUUAUUUUGCUUUUAUGAUGACUGACAUAUCACUGCAUGACACAUUUUCCCUCUAUUUGAGUUUUUAGCUCAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAA- ...................((((((...((((......(((.((.........((..(((((.....)))))..))..)).)))......))))...))))))......- ( -13.90) >DroGri_CAF1 34165 109 - 1 UUUUU-UUUCUUUUAUGAUGACUGACAUAUCACUGCAUGACACAUUUUCCCUCUAUUUGAGUUUUUAGCUCAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAAA .....-.....(((((...((((((...((((......(((.((.........((..(((((.....)))))..))..)).)))......))))...))))))..))))) ( -14.60) >DroAna_CAF1 22590 108 - 1 UUUCG-CUUUUAUUUUGUUGACUGACAUAUCACUGCAUGACACAUUUUCCCUCUAUUUGAGUUUUUAGCUCAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAA- .....-.............((((((...((((......(((.((.........((..(((((.....)))))..))..)).)))......))))...))))))......- ( -13.90) >DroPer_CAF1 26284 109 - 1 UUUAUUUUGCUUUUAUGAUGACUGACAUAUCACUGCAUGACACAUUUUCCCUCUAUUUGAGUUUUUAGCUCAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAA- ...................((((((...((((......(((.((.........((..(((((.....)))))..))..)).)))......))))...))))))......- ( -13.90) >consensus UUUAU_UUGCUUUUAUGAUGACUGACAUAUCACUGCAUGACACAUUUUCCCUCUAUUUGAGUUUUUAGCUCAUUUACUUGCGUUUCUAUUUGAUUUUUCAGUUCUGUAA_ ...................((((((...((((......(((.((.........((..(((((.....)))))..))..)).)))......))))...))))))....... (-13.90 = -13.90 + 0.00)


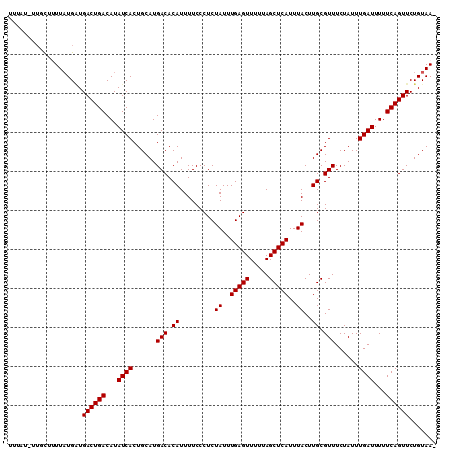
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:06 2006