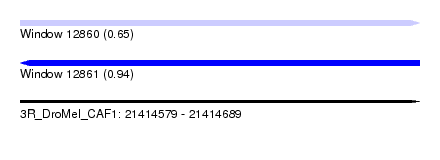
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,414,579 – 21,414,689 |
| Length | 110 |
| Max. P | 0.941243 |

| Location | 21,414,579 – 21,414,689 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.76 |
| Mean single sequence MFE | -24.45 |
| Consensus MFE | -19.13 |
| Energy contribution | -19.47 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21414579 110 + 27905053 UUGCUAAUGACGCUCAACAAUGUUAAUGCCCCGACCCAGCAGUAAACUA--UAAUAUAGUGUUACAAUACUGCUGUGACAGGACAAUAUCAAUGGCAAGCGUCC-GAUUUCAU------- ........((((((...((.((.((.((.((.(.(.((((((((.((((--(...))))).......)))))))).).).)).)).)).)).))...)))))).-........------- ( -26.30) >DroVir_CAF1 9910 108 + 1 UUGCUAACGACGCUCAACAAUGUUAAUGCCCCGGCUCAGCAGUACCAAU--UGCUUA-A-CGUACAAUACUGCUGUGACAGGACAAUAUCACUGGCAAGCGCAACGAUCGA-C------- ........(.((((...((.((.((.((.((.(.(.((((((((.....--(((...-.-.)))...)))))))).).).)).)).)).)).))...))))).........-.------- ( -23.40) >DroWil_CAF1 115104 111 + 1 UUGCCACAGACGCUCAACAAUGUUAAUGCCCCGGCUCAGCAGUACGAAA--UGAAUA--UGUUACUAUACUGCUGUGACAGGACAAUAUCACUGGCAAGCUCUC---UUUUGUUAGAU-- (((((...(.....).....(((((..((....)).((((((((...((--(.....--.)))....))))))))))))))).))).....((((((((.....---.))))))))..-- ( -22.30) >DroMoj_CAF1 9521 108 + 1 UUGCUAACGACGCUCAACAAUGUUAAUGCCCCGGCUCAGCAGUACCAAU--UGCUUA-A-UUUACAAUACUGCUGUGACAGGACAAUAUCACUGGCAAGCGCUAUAAUAGA-C------- ........(.((((...((.((.((.((.((.(.(.((((((((....(--((....-.-....))))))))))).).).)).)).)).)).))...))))).........-.------- ( -21.40) >DroAna_CAF1 100978 103 + 1 UUGCUAAUGACGCUCAACAAUGUUAAUGCCCCGACCCAGCAGUAAGCUAUAUAGUAUAGAAUUACAAUACUGCUGUGACAGGACAAUAUCAUCGGCAAGCGUC----------------- ........((((((...(.(((.((.((.((.(.(.((((((((..(((((...)))))........)))))))).).).)).)).)).))).)...))))))----------------- ( -24.70) >DroPer_CAF1 8945 113 + 1 UUGCCAAUGGCGCUCAACAAUGUUAAUGCCCCGACCCAGCAGUAGGCUA--UAAUUAA--GUUACAAUACUGCUGUGACAGGACAAUAUCAAUGGCAAGCGCCC---UCUCAUCUGCUCC ........((((((...((.((.((.((.((.(.(.((((((((((((.--......)--)))....)))))))).).).)).)).)).)).))...)))))).---............. ( -28.60) >consensus UUGCUAAUGACGCUCAACAAUGUUAAUGCCCCGACCCAGCAGUACGAAA__UAAUUA___GUUACAAUACUGCUGUGACAGGACAAUAUCACUGGCAAGCGCCC___UCUA_U_______ ..........((((...((.((.((.((.((.(.(.((((((((.......................)))))))).).).)).)).)).)).))...))))................... (-19.13 = -19.47 + 0.33)

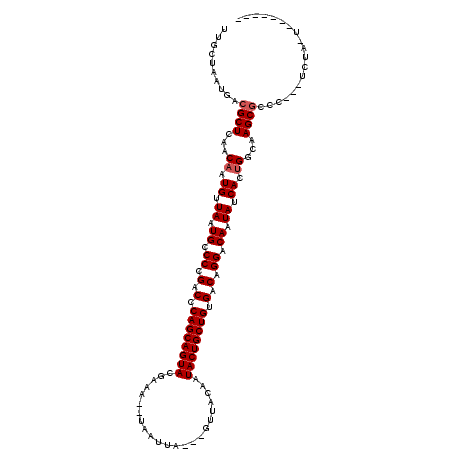
| Location | 21,414,579 – 21,414,689 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.76 |
| Mean single sequence MFE | -41.51 |
| Consensus MFE | -36.43 |
| Energy contribution | -36.35 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.29 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21414579 110 - 27905053 -------AUGAAAUC-GGACGCUUGCCAUUGAUAUUGUCCUGUCACAGCAGUAUUGUAACACUAUAUUA--UAGUUUACUGCUGGGUCGGGGCAUUAACAUUGUUGAGCGUCAUUAGCAA -------.((.....-.(((((((..((.((.((.(((((((.(.((((((((((((((.......)))--))))..))))))).).))))))).)).)).))..))))))).....)). ( -41.40) >DroVir_CAF1 9910 108 - 1 -------G-UCGAUCGUUGCGCUUGCCAGUGAUAUUGUCCUGUCACAGCAGUAUUGUACG-U-UAAGCA--AUUGGUACUGCUGAGCCGGGGCAUUAACAUUGUUGAGCGUCGUUAGCAA -------(-(..(.((..((((((..(((((.((.(((((((.(.(((((((((((...(-.-....).--..))))))))))).).))))))).)).)))))..)))))))))..)).. ( -42.30) >DroWil_CAF1 115104 111 - 1 --AUCUAACAAAA---GAGAGCUUGCCAGUGAUAUUGUCCUGUCACAGCAGUAUAGUAACA--UAUUCA--UUUCGUACUGCUGAGCCGGGGCAUUAACAUUGUUGAGCGUCUGUGGCAA --..........(---.(((((((..(((((.((.(((((((.(.(((((((((.......--......--....))))))))).).))))))).)).)))))..)))).))).)..... ( -37.33) >DroMoj_CAF1 9521 108 - 1 -------G-UCUAUUAUAGCGCUUGCCAGUGAUAUUGUCCUGUCACAGCAGUAUUGUAAA-U-UAAGCA--AUUGGUACUGCUGAGCCGGGGCAUUAACAUUGUUGAGCGUCGUUAGCAA -------(-.(((..(..((((((..(((((.((.(((((((.(.((((((((((((...-.-...)))--....))))))))).).))))))).)).)))))..))))))..))))).. ( -40.90) >DroAna_CAF1 100978 103 - 1 -----------------GACGCUUGCCGAUGAUAUUGUCCUGUCACAGCAGUAUUGUAAUUCUAUACUAUAUAGCUUACUGCUGGGUCGGGGCAUUAACAUUGUUGAGCGUCAUUAGCAA -----------------(((((((..(((((.((.(((((((.(.(((((((((((((...........)))))..)))))))).).))))))).)).)))))..)))))))........ ( -42.70) >DroPer_CAF1 8945 113 - 1 GGAGCAGAUGAGA---GGGCGCUUGCCAUUGAUAUUGUCCUGUCACAGCAGUAUUGUAAC--UUAAUUA--UAGCCUACUGCUGGGUCGGGGCAUUAACAUUGUUGAGCGCCAUUGGCAA ...((....((..---.(((((((..((.((.((.(((((((.(.((((((((((((((.--....)))--)))..)))))))).).))))))).)).)).))..))))))).)).)).. ( -44.40) >consensus _______A_GAAA___GGGCGCUUGCCAGUGAUAUUGUCCUGUCACAGCAGUAUUGUAAC___UAAUCA__UAGCGUACUGCUGAGCCGGGGCAUUAACAUUGUUGAGCGUCAUUAGCAA ..................((((((..(((((.((.(((((((.(.((((((((.......................)))))))).).))))))).)).)))))..))))))......... (-36.43 = -36.35 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:41 2006