
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,392,550 – 21,392,671 |
| Length | 121 |
| Max. P | 0.994646 |

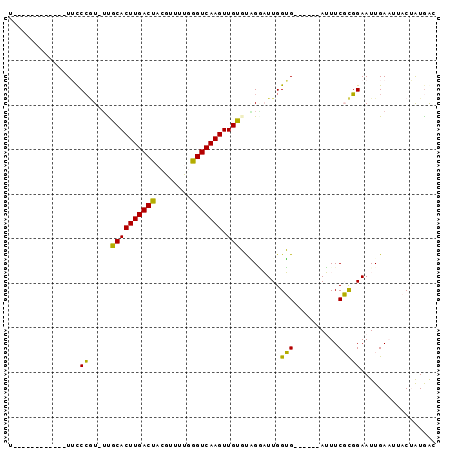
| Location | 21,392,550 – 21,392,640 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 68.86 |
| Mean single sequence MFE | -24.56 |
| Consensus MFE | -13.14 |
| Energy contribution | -11.62 |
| Covariance contribution | -1.52 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21392550 90 + 27905053 UCCUCUGUAGCAUUGCCCGUUUUGCACUUGACUACGUUUUGGGUCAAGUUGCGUAGAAUUGGUG------AUUUCGCGGAAUUGAAUUACUAUGAC ...((((..(.((..((.(((((((((((((((........))))))))...))))))).))..------)).)..))))................ ( -26.30) >DroPse_CAF1 106862 79 + 1 U------------UUUCCAU-GUGCACUUGACUCAAUUUUGAGUCAAGUUGUUUCGUUUUGGGGCAAGCCAAUUCCUGGAAUUGA----CAGUGAC .------------.((((((-(.((((((((((((....))))))))))(((..(.....)..))).)))).....)))))....----....... ( -23.50) >DroSim_CAF1 83333 77 + 1 U------------UGCCCGU-UUGCACUUGACUACGUUUUGGGUCAAGUUGCGCGGGAUUGGUG------AUUUCGCGGAAUUGAAUUACUAUAAC .------------..(((((-..((((((((((........))))))).))))))))..(((((------((((.........))))))))).... ( -25.00) >DroYak_CAF1 86605 89 + 1 UUCUCUGUAGCGUUUCCAGU-UCGCACUUGACUACGUUUUGGGUCAAGUUGUGUAGGAUUGGUG------AUUUCGCUGAAUUGAAUUACUAUGGC (((....(((((...(((((-((((((((((((........))))))).)))...)))))))..------....)))))....))).......... ( -24.90) >DroPer_CAF1 129784 79 + 1 U------------UUUCCCU-UUGCACUUGACUCAAUUUUGAGUCAAGUUGUUUCGUUUUGGGGCAAGCCAAUUCCUGGAAUUGA----CAGUGAC .------------.......-(..(((((((((((....))))))))))((((..((((..(((.........)))..)))).))----)))..). ( -23.10) >consensus U____________UUCCCGU_UUGCACUUGACUACGUUUUGGGUCAAGUUGUGUAGGAUUGGUG______AUUUCGCGGAAUUGAAUUACUAUGAC ................((.....((((((((((........))))))).))).........(((..........)))))................. (-13.14 = -11.62 + -1.52)


| Location | 21,392,550 – 21,392,640 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 68.86 |
| Mean single sequence MFE | -17.32 |
| Consensus MFE | -7.44 |
| Energy contribution | -6.84 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21392550 90 - 27905053 GUCAUAGUAAUUCAAUUCCGCGAAAU------CACCAAUUCUACGCAACUUGACCCAAAACGUAGUCAAGUGCAAAACGGGCAAUGCUACAGAGGA ....(((((.......((((.(((..------......)))...(((.((((((..........)))))))))....))))...)))))....... ( -14.40) >DroPse_CAF1 106862 79 - 1 GUCACUG----UCAAUUCCAGGAAUUGGCUUGCCCCAAAACGAAACAACUUGACUCAAAAUUGAGUCAAGUGCAC-AUGGAAA------------A .......----....(((((....((((......)))).........((((((((((....))))))))))....-.))))).------------. ( -21.20) >DroSim_CAF1 83333 77 - 1 GUUAUAGUAAUUCAAUUCCGCGAAAU------CACCAAUCCCGCGCAACUUGACCCAAAACGUAGUCAAGUGCAA-ACGGGCA------------A ................((((((....------.........)))(((.((((((..........)))))))))..-..)))..------------. ( -15.02) >DroYak_CAF1 86605 89 - 1 GCCAUAGUAAUUCAAUUCAGCGAAAU------CACCAAUCCUACACAACUUGACCCAAAACGUAGUCAAGUGCGA-ACUGGAAACGCUACAGAGAA ....((((..((((.(((.(((....------)..............(((((((..........)))))))))))-).))))...))))....... ( -13.50) >DroPer_CAF1 129784 79 - 1 GUCACUG----UCAAUUCCAGGAAUUGGCUUGCCCCAAAACGAAACAACUUGACUCAAAAUUGAGUCAAGUGCAA-AGGGAAA------------A ......(----(((((((...))))))))....(((...........((((((((((....))))))))))....-.)))...------------. ( -22.49) >consensus GUCAUAGUAAUUCAAUUCCGCGAAAU______CACCAAUACGACACAACUUGACCCAAAACGUAGUCAAGUGCAA_ACGGGAA____________A ................((((.(.............)...........(((((((..........)))))))......))))............... ( -7.44 = -6.84 + -0.60)



| Location | 21,392,574 – 21,392,671 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 69.37 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -7.18 |
| Energy contribution | -7.18 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21392574 97 - 27905053 UAAAGUCAACAGAGUGGUAAAUUCA---------GCCUCCGUCAUAGUAAUUCAAUUCCGCGAAAU------CACCAAUUCUACGCAACUUGACCCAAAACGUAGUCAAGUG ..........((((((((.(.(((.---------((.......................))))).)------.))).))))).....(((((((..........))))))). ( -13.10) >DroPse_CAF1 106873 108 - 1 UGUAGCCACCGGAGGAGUAAAUUUAUCUCUUCUGGCCACUGUCACUG----UCAAUUCCAGGAAUUGGCUUGCCCCAAAACGAAACAACUUGACUCAAAAUUGAGUCAAGUG ((..((..(((((((((.........)))))))))...........(----(((((((...))))))))..))..))..........((((((((((....)))))))))). ( -35.00) >DroSim_CAF1 83344 97 - 1 UAAAGUCAACAGAGUGGUAAAUUCA---------GCCUCUGUUAUAGUAAUUCAAUUCCGCGAAAU------CACCAAUCCCGCGCAACUUGACCCAAAACGUAGUCAAGUG .......((((((((((.....)))---------..)))))))...............((((....------.........))))..(((((((..........))))))). ( -18.62) >DroYak_CAF1 86628 85 - 1 ------------AGUGGUAAAUUCU---------GCCUCCGCCAUAGUAAUUCAAUUCAGCGAAAU------CACCAAUCCUACACAACUUGACCCAAAACGUAGUCAAGUG ------------.(.((((.....)---------))).)(((...(((......)))..)))....------...............(((((((..........))))))). ( -11.20) >DroPer_CAF1 129795 108 - 1 UGUAGCAACCAGAGGAGUAAAUUUAUCUCUUCUGGCCACUGUCACUG----UCAAUUCCAGGAAUUGGCUUGCCCCAAAACGAAACAACUUGACUCAAAAUUGAGUCAAGUG ((..(((((((((((((.........)))))))))...........(----(((((((...))))))))))))..))..........((((((((((....)))))))))). ( -38.00) >consensus UAAAGCCAACAGAGUGGUAAAUUCA_________GCCUCUGUCAUAGUAAUUCAAUUCCGCGAAAU______CACCAAUACGACACAACUUGACCCAAAACGUAGUCAAGUG .......................................................................................(((((((..........))))))). ( -7.18 = -7.18 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:29 2006