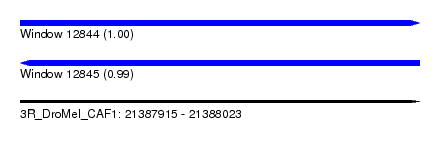
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,387,915 – 21,388,023 |
| Length | 108 |
| Max. P | 0.997012 |

| Location | 21,387,915 – 21,388,023 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.15 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -12.48 |
| Energy contribution | -16.12 |
| Covariance contribution | 3.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.40 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.997012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
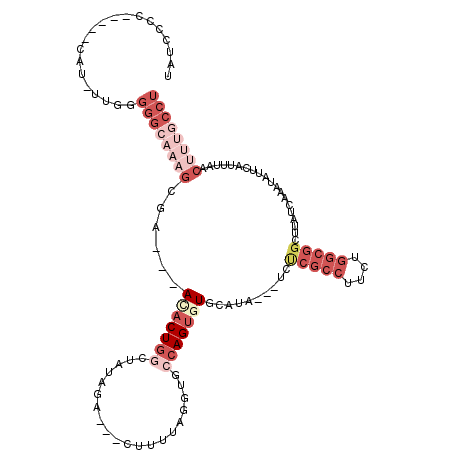
>3R_DroMel_CAF1 21387915 108 + 27905053 UAUCCCC-----CAU-UUGGGGGCAAAGCGA---ACACUGGCUAUAGA---CUUUUAGGUGCCAGUGUGCAUAAUAUCUCGCCUUCUGGCGGCUUAUCAAAUAUUCACUUAACUUUGCCA .....((-----(..-..)))(((((((.(.---((((((((..(((.---...)))...)))))))).)...(((..(((((....)))))..)))...............))))))). ( -36.70) >DroSec_CAF1 82121 107 + 1 UAUGCCC-----CAU-UUGGGGGCA-AGCGA---ACACUGGCUAUAGA---CUUUUAUGUGCCAGUGUACAUAAUAUCCCGCCUUCUGGCGGCUUAUCAAAUAUUCAUUUAACUUUGCCU ..(((((-----(..-...))))))-.((((---(((((((((((((.---...))))).)))))))).....(((..(((((....)))))..))).................)))).. ( -40.00) >DroSim_CAF1 78669 108 + 1 UAUGCCC-----CAU-UUGGGGGCAAAGCGA---ACACUGGCUAUAGA---CUUUUAGUUGCCAGUGUACAUAAUAUCUCGCCUUCUGGCGGCUUAUCAAAUAUUCAUUUAACUUUGCCU ..(((((-----(..-...))))))(((((.---((((((((....((---(.....))))))))))).).........((((....))))))))......................... ( -36.50) >DroEre_CAF1 81995 97 + 1 UAUUCCCAUAUCCAUUUUUAGGG------GAUAUAGACU--------------UUUGGGUGCCAGUGUGCAUA---UCUCGCCUUCUGGCGGCUUAUCAAAUAUUCAUUUAACUUUGCCU (((((((.((........)))))------))))..((..--------------(((((..(((.(....)...---....(((....))))))...)))))...)).............. ( -20.20) >DroYak_CAF1 81752 116 + 1 UAUCCCCCC-CACAUUUUGGGGGCAAUGAGAUAUAGACUGGAUAUAGAUGAUAUUUGGGUGCCAGUGUGCAUA---ACUCGCCUUCUGGCGGCUUAUCACUUAUUCAUUUAACUUUGCCU .....((((-((.....)))))).((((((......(((((...(((((...)))))....)))))(((.(((---(..((((....))))..)))))))...))))))........... ( -33.30) >DroPer_CAF1 123858 89 + 1 UAUCC-G-----CAU-UUAGG--AAAAGAGC---AUUCUGAAUGGAUG---CUUUUGGCAGGCAGUGU----------------UUUGGCGGCUUAUCAAAUAAUCAUUUAACUUCGUUU ..(((-.-----...-...))--).((((((---((((.....)))))---)))))(..((((.((..----------------....)).))))..)...................... ( -19.20) >consensus UAUCCCC_____CAU_UUGGGGGCAAAGCGA___ACACUGGCUAUAGA___CUUUUAGGUGCCAGUGUGCAUA___UCUCGCCUUCUGGCGGCUUAUCAAAUAUUCAUUUAACUUUGCCU ....................((((((((......(((((((....................)))))))..........(((((....)))))....................)))))))) (-12.48 = -16.12 + 3.64)

| Location | 21,387,915 – 21,388,023 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.15 |
| Mean single sequence MFE | -29.36 |
| Consensus MFE | -12.36 |
| Energy contribution | -13.59 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.42 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
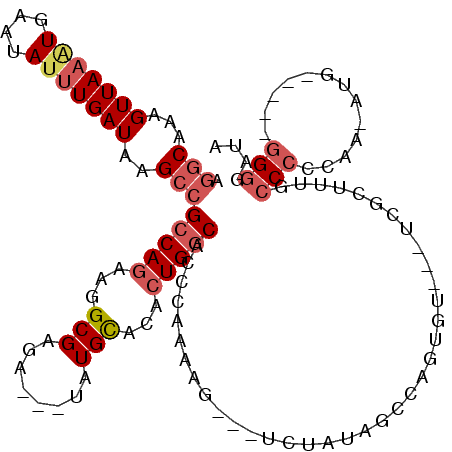
>3R_DroMel_CAF1 21387915 108 - 27905053 UGGCAAAGUUAAGUGAAUAUUUGAUAAGCCGCCAGAAGGCGAGAUAUUAUGCACACUGGCACCUAAAAG---UCUAUAGCCAGUGU---UCGCUUUGCCCCCAA-AUG-----GGGGAUA .(((...(((((((....)))))))..))).....(((((((..........((((((((.........---......))))))))---))))))).(((((..-..)-----))))... ( -35.76) >DroSec_CAF1 82121 107 - 1 AGGCAAAGUUAAAUGAAUAUUUGAUAAGCCGCCAGAAGGCGGGAUAUUAUGUACACUGGCACAUAAAAG---UCUAUAGCCAGUGU---UCGCU-UGCCCCCAA-AUG-----GGGCAUA ((((.................(((((..(((((....)))))..))))).(.((((((((.........---......))))))))---.))))-)(((((...-..)-----))))... ( -37.96) >DroSim_CAF1 78669 108 - 1 AGGCAAAGUUAAAUGAAUAUUUGAUAAGCCGCCAGAAGGCGAGAUAUUAUGUACACUGGCAACUAAAAG---UCUAUAGCCAGUGU---UCGCUUUGCCCCCAA-AUG-----GGGCAUA .((((((((............(((((...((((....))))...))))).(.((((((((.........---......))))))))---.)))))))))(((..-..)-----))..... ( -35.76) >DroEre_CAF1 81995 97 - 1 AGGCAAAGUUAAAUGAAUAUUUGAUAAGCCGCCAGAAGGCGAGA---UAUGCACACUGGCACCCAAA--------------AGUCUAUAUC------CCCUAAAAAUGGAUAUGGGAAUA .(((...(((((((....)))))))..)))(((((...(((...---..)))...))))).......--------------..((((((((------(.........))))))))).... ( -26.50) >DroYak_CAF1 81752 116 - 1 AGGCAAAGUUAAAUGAAUAAGUGAUAAGCCGCCAGAAGGCGAGU---UAUGCACACUGGCACCCAAAUAUCAUCUAUAUCCAGUCUAUAUCUCAUUGCCCCCAAAAUGUG-GGGGGGAUA ((((.....((.((((....(((...(((((((....)))).))---)...)))..(((...)))....)))).))......)))).......(((.((((((.....))-)))).))). ( -30.60) >DroPer_CAF1 123858 89 - 1 AAACGAAGUUAAAUGAUUAUUUGAUAAGCCGCCAAA----------------ACACUGCCUGCCAAAAG---CAUCCAUUCAGAAU---GCUCUUUU--CCUAA-AUG-----C-GGAUA .......(((((((....)))))))...((((((..----------------....)).........((---(((.(.....).))---))).....--.....-..)-----)-))... ( -9.60) >consensus AGGCAAAGUUAAAUGAAUAUUUGAUAAGCCGCCAGAAGGCGAGA___UAUGCACACUGGCACCCAAAAG___UCUAUAGCCAGUGU___UCGCUUUGCCCCCAA_AUG_____GGGGAUA .(((...(((((((....)))))))..)))(((((...(((........)))...))))).....................................(((.............))).... (-12.36 = -13.59 + 1.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:27 2006