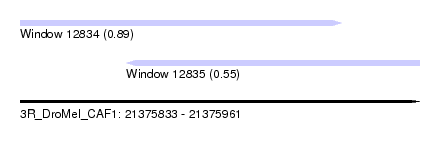
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,375,833 – 21,375,961 |
| Length | 128 |
| Max. P | 0.890660 |

| Location | 21,375,833 – 21,375,936 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 93.07 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -20.70 |
| Energy contribution | -21.14 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
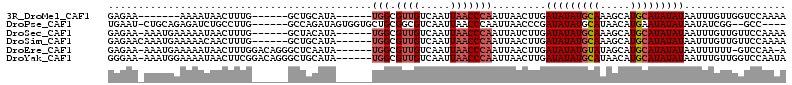
>3R_DroMel_CAF1 21375833 103 + 27905053 GAGCCCAGCUCUGAUUGUUGUUAUGGAGUUGGCAUUUUGGACCAACAAAUUAUAUAUGCAUGCUUUGCAUAUAUCAAGUUAAUUGGGUUAAUUGACAACGCCA ..(((.(((((((((....)))..)))))))))....(((.((((..(((((((((((((.....)))))))))..))))..))))(((....)))....))) ( -26.30) >DroSec_CAF1 70341 102 + 1 GAC-CCAGCUCUGAUUGUUGUUAUGGAGUUGCCAUUUUGGAACAACAAAUUAUAUAUGCAUGCUUUGCAUAUAUCAAGAUAAUUGGGUUAAUUGACAACGCCA (((-((((.(((((((((((((((((.....)))).....))))))))....((((((((.....)))))))))).)))...))))))).............. ( -27.30) >DroSim_CAF1 66717 102 + 1 GAG-CCGGCUCUGAUUGUUGUUAUGGAGUUGGCAUUUUGGAACAACAAAUUAUAUAUGCAUGCUUUGCAUAUAUCAAGUUAAUUGGGUUAAUUGACAACGCCA ..(-(((((((((((....)))..)))))))))....(((...........(((((((((.....)))))))))...((((((((...))))))))....))) ( -28.40) >DroEre_CAF1 69783 100 + 1 GAG-CCAGCUCUGAUUCUUGUUAUGGAGUUGGCUU-UUGGAC-AAAAAAUUAUAUAUGCAUGCUAUACAUAUAUCAAGUUAAUUGGGUUAAUUGACAACGCCA (((-(((((((((((....)))..)))))))))))-.(((.(-........(((((((.........)))))))...((((((((...))))))))...)))) ( -24.30) >DroYak_CAF1 69280 102 + 1 GAG-CCAGCUCUGAUUCUUCUUAUGGAGUUGGCUUAUUGGACCAACAAAUUAUAUAUGCAUGUUAUGCAUAUAUCAAGUUAAUUGGGUUAAUUGACAACGCCA (((-((((((((............)))))))))))..(((.((((..((((((((((((((...))))))))))..))))..))))(((....)))....))) ( -31.10) >consensus GAG_CCAGCUCUGAUUGUUGUUAUGGAGUUGGCAUUUUGGACCAACAAAUUAUAUAUGCAUGCUUUGCAUAUAUCAAGUUAAUUGGGUUAAUUGACAACGCCA ....((((((((............)))))))).....(((...........(((((((((.....)))))))))...((((((((...))))))))....))) (-20.70 = -21.14 + 0.44)

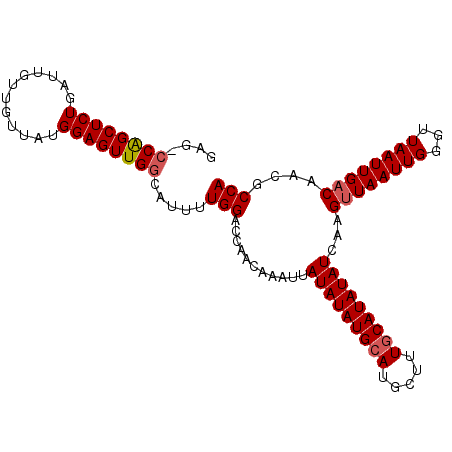
| Location | 21,375,867 – 21,375,961 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.25 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -9.94 |
| Energy contribution | -10.63 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21375867 94 - 27905053 GAGAA-------AAAAUAACUUUG------GCUGCAUA------UGGCGUUGUCAAUUAACCCAAUUAACUUGAUAUAUGCAAAGCAUGCAUAUAUAAUUUGUUGGUCCAAAA .....-------........((((------(.......------(((.((((.....)))))))((((((...(((((((((.....))))))))).....))))))))))). ( -20.50) >DroPse_CAF1 85712 100 - 1 UGAAU-CUGCAGAGAUCUGCCUUG------GCCAGAUAGUGGUGCUGCGGCGUCAAUUAACCCAAUUAACCCGAUAUAUGCAUAACAUGAAUAUAUAAUAUCGG--GCC---- (((..-((((((..((((((....------).))))).......))))))..)))..............(((((((((((.((.......)).))).)))))))--)..---- ( -24.80) >DroSec_CAF1 70374 100 - 1 GAGAA-AAAUGAAAAAUAACUUUG------GCUACAUA------UGGCGUUGUCAAUUAACCCAAUUAUCUUGAUAUAUGCAAAGCAUGCAUAUAUAAUUUGUUGUUCCAAAA .....-..............((((------(..(((..------(((.((((.....))))))).........(((((((((.....))))))))).......))).))))). ( -19.40) >DroSim_CAF1 66750 101 - 1 GAGAACAAAUGAAAAACAACUUUG------GCUGCAUA------UGGCGUUGUCAAUUAACCCAAUUAACUUGAUAUAUGCAAAGCAUGCAUAUAUAAUUUGUUGUUCCAAAA ....................((((------(..(((..------(((.((((.....))))))).........(((((((((.....))))))))).......))).))))). ( -19.70) >DroEre_CAF1 69816 104 - 1 GAGAA-AAAUGAAAAAUAACUUUGGACAGGGCUCAAUA------UGGCGUUGUCAAUUAACCCAAUUAACUUGAUAUAUGUAUAGCAUGCAUAUAUAAUUUUUU-GUCCAA-A .....-...............((((((((((.......------(((.((((.....))))))).........((((((((((...))))))))))....))))-))))))-. ( -22.80) >DroYak_CAF1 69313 106 - 1 GGGAA-AAAUGGAAAAUAACUUCGGACAGGGCUGCAUA------UGGCGUUGUCAAUUAACCCAAUUAACUUGAUAUAUGCAUAACAUGCAUAUAUAAUUUGUUGGUCCAAUA .....-...((((...........(((((.(((.....------.))).))))).......(((((.......((((((((((...)))))))))).....)))))))))... ( -28.80) >consensus GAGAA_AAAUGAAAAAUAACUUUG______GCUGCAUA______UGGCGUUGUCAAUUAACCCAAUUAACUUGAUAUAUGCAAAGCAUGCAUAUAUAAUUUGUUGGUCCAAAA ............................................(((.((((.....))))))).........(((((((((.....)))))))))................. ( -9.94 = -10.63 + 0.69)
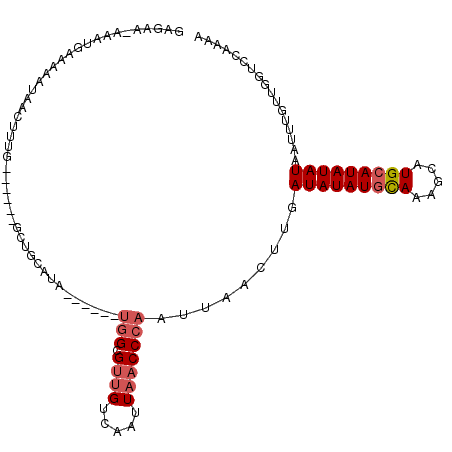
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:17 2006