
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,362,571 – 21,362,711 |
| Length | 140 |
| Max. P | 0.967003 |

| Location | 21,362,571 – 21,362,671 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.93 |
| Mean single sequence MFE | -29.89 |
| Consensus MFE | -19.58 |
| Energy contribution | -20.75 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.967003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21362571 100 + 27905053 CAUCAGCUGUCAGCAGCUUU------GGCUGCUGUCUGCCUAAAUAUUUGCCAGCUGCGUAAUUGCAGCCGAAACUACGGAGCAAAAAA-----UAAGACAGAGUCGACAA ....(((((....)))))((------((((.((((((.........(((((..((((((....)))))).....(....).)))))...-----..))))))))))))... ( -32.44) >DroPse_CAF1 66744 89 + 1 CAUCAACUGUCAACAGCCUUUGCUUCUGCUGCUGUCUGCGUAAAUAUUUGCCAGCUGCGUAAUUGCAGCUGAAACCG------AAAAAA----------------CUACAG ......((((..(((((....((....)).)))))..(((........)))((((((((....))))))))......------......----------------..)))) ( -23.20) >DroSim_CAF1 52415 99 + 1 CAUCAGCUGUCAGCAGCUUU------GGCUGCUGUCUGCCUAAAUAUUUGCCAGCUGCGUAAUUGCAGCCGAAACUGCGGAGCAAAAAA-----UAAGACAGUGUCGACA- .....((((((((((((...------.)))))).............((((((.((((((....))))))((......))).)))))...-----...)))))).......- ( -32.20) >DroEre_CAF1 54817 105 + 1 CAUCAGCUGUCAGCAGCUUU------GGCUGCUGUCUGCCUAAAUAUUUGCCAGCUGCGUAAUUGCAGGCGAAACUAGGGAGGUAAAAAUAAAAAGAGCUAGUGUCUACAA (((.((((..(((((((...------.)))))))(((.((((....((((((...((((....))))))))))..)))).))).............)))).)))....... ( -34.30) >DroYak_CAF1 55080 101 + 1 CAUCAGCUGUCAGCAGCUUU------GGCUGCUGUCUGCCUAAAUAUUUGCCAGCUGCGUAAUUGCAGCCGAAACUGCGGUGGAAAAAA----AUAAGACAGUGUCUACAA .....((((((((((((...------.)))))).((..((......((((...((((((....)))))))))).....))..)).....----....))))))........ ( -34.00) >DroPer_CAF1 88644 89 + 1 CAUCAACUGUCAACAGCCUUUGCUUCUGCUGCUGUCUGCGUAAAUAUUUGCCAGCUGCGUAAUUGCAGCUGAAACCG------AAAAAA----------------CUACAG ......((((..(((((....((....)).)))))..(((........)))((((((((....))))))))......------......----------------..)))) ( -23.20) >consensus CAUCAGCUGUCAGCAGCUUU______GGCUGCUGUCUGCCUAAAUAUUUGCCAGCUGCGUAAUUGCAGCCGAAACUGCGGAG_AAAAAA_____UAAGACAGUGUCUACAA ..((.((.(.((((((((........)))))))).).))..............((((((....)))))).))....................................... (-19.58 = -20.75 + 1.17)



| Location | 21,362,571 – 21,362,671 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.93 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -24.34 |
| Energy contribution | -23.40 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21362571 100 - 27905053 UUGUCGACUCUGUCUUA-----UUUUUUGCUCCGUAGUUUCGGCUGCAAUUACGCAGCUGGCAAAUAUUUAGGCAGACAGCAGCC------AAAGCUGCUGACAGCUGAUG ..((((..(((((((..-----...((((((.((......))(((((......))))).)))))).....)))))))(((((((.------...))))))).....)))). ( -36.90) >DroPse_CAF1 66744 89 - 1 CUGUAG----------------UUUUUU------CGGUUUCAGCUGCAAUUACGCAGCUGGCAAAUAUUUACGCAGACAGCAGCAGAAGCAAAGGCUGUUGACAGUUGAUG ((((.(----------------(.....------.....((((((((......)))))))).........)))))).(((((((..........))))))).......... ( -25.31) >DroSim_CAF1 52415 99 - 1 -UGUCGACACUGUCUUA-----UUUUUUGCUCCGCAGUUUCGGCUGCAAUUACGCAGCUGGCAAAUAUUUAGGCAGACAGCAGCC------AAAGCUGCUGACAGCUGAUG -.((((.(.((((((..-----...(((((..........(((((((......)))))))))))).....)))))).(((((((.------...)))))))...).)))). ( -36.40) >DroEre_CAF1 54817 105 - 1 UUGUAGACACUAGCUCUUUUUAUUUUUACCUCCCUAGUUUCGCCUGCAAUUACGCAGCUGGCAAAUAUUUAGGCAGACAGCAGCC------AAAGCUGCUGACAGCUGAUG .......((.(((((..............((.(((((.((.(((.((.........)).))).))...))))).)).(((((((.------...)))))))..))))).)) ( -28.60) >DroYak_CAF1 55080 101 - 1 UUGUAGACACUGUCUUAU----UUUUUUCCACCGCAGUUUCGGCUGCAAUUACGCAGCUGGCAAAUAUUUAGGCAGACAGCAGCC------AAAGCUGCUGACAGCUGAUG ((((.....((((((...----..(((.(((.((......))(((((......)))))))).))).....)))))).(((((((.------...)))))))))))...... ( -31.50) >DroPer_CAF1 88644 89 - 1 CUGUAG----------------UUUUUU------CGGUUUCAGCUGCAAUUACGCAGCUGGCAAAUAUUUACGCAGACAGCAGCAGAAGCAAAGGCUGUUGACAGUUGAUG ((((.(----------------(.....------.....((((((((......)))))))).........)))))).(((((((..........))))))).......... ( -25.31) >consensus UUGUAGACACUGUCUUA_____UUUUUU_CUCCGCAGUUUCGGCUGCAAUUACGCAGCUGGCAAAUAUUUAGGCAGACAGCAGCC______AAAGCUGCUGACAGCUGAUG ..................................(((((((((((((......))))))))................(((((((..........)))))))..)))))... (-24.34 = -23.40 + -0.94)

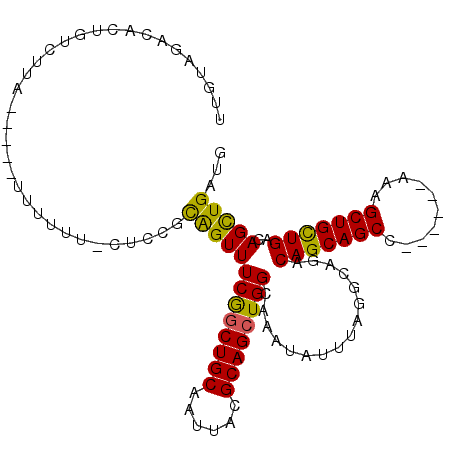

| Location | 21,362,596 – 21,362,711 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.94 |
| Mean single sequence MFE | -23.68 |
| Consensus MFE | -19.39 |
| Energy contribution | -19.87 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21362596 115 - 27905053 AAAUAUAUGUUUGUUUUAGCCUCUUUCUUUUUUUUUUUUUUUGUCGACUCUGUCUUA-----UUUUUUGCUCCGUAGUUUCGGCUGCAAUUACGCAGCUGGCAAAUAUUUAGGCAGACAG ................................................(((((((..-----...((((((.((......))(((((......))))).)))))).....)))))))... ( -23.30) >DroSec_CAF1 55366 110 - 1 AAAUAUAUGUUUGUUUUAGCCUCAUU-----UUUUUUUUUUCGUCGACACUGUCUUA-----UUUUUUGCUCCGUAGUUUCGGCUGCAAUUACGCAGCUGGCAAAUAUUUAGGCAGACAG ..........................-----..................((((((..-----...((((((.((......))(((((......))))).)))))).....)))))).... ( -22.40) >DroSim_CAF1 52440 105 - 1 GAAUAUAUGUUUGUUUUAGCCUCAUU-----UUUUU-----UGUCGACACUGUCUUA-----UUUUUUGCUCCGCAGUUUCGGCUGCAAUUACGCAGCUGGCAAAUAUUUAGGCAGACAG ...........(((((..((((....-----...((-----((((((.(((((....-----...........))))).))((((((......)))))))))))).....))))))))). ( -27.26) >DroEre_CAF1 54842 107 - 1 AAAUAUAUGUAUGUUUUAGCCUCUUU-------------UUUGUAGACACUAGCUCUUUUUAUUUUUACCUCCCUAGUUUCGCCUGCAAUUACGCAGCUGGCAAAUAUUUAGGCAGACAG ...........(((((..((((....-------------(((((.((.(((((....................))))).))((.(((......)))))..))))).....))))))))). ( -19.65) >DroYak_CAF1 55105 104 - 1 ACAUUUAUGUGUGUGCUAGCCGCUUU------------UUUUGUAGACACUGUCUUAU----UUUUUUCCACCGCAGUUUCGGCUGCAAUUACGCAGCUGGCAAAUAUUUAGGCAGACAG ((((....))))((.((.(((.....------------.(((((.((.(((((.....----...........))))).))((((((......)))))).)))))......))))))).. ( -25.79) >consensus AAAUAUAUGUUUGUUUUAGCCUCUUU_____UUUUU__UUUUGUCGACACUGUCUUA_____UUUUUUGCUCCGUAGUUUCGGCUGCAAUUACGCAGCUGGCAAAUAUUUAGGCAGACAG .......(((((((((..(((........................((.(((((....................))))).))((((((......)))))))))........))))))))). (-19.39 = -19.87 + 0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:59:11 2006