| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
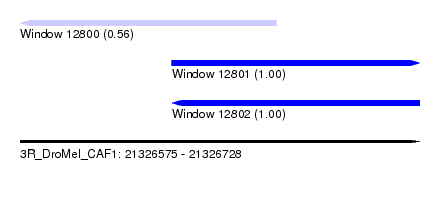
| Location | 21,326,575 – 21,326,728 |
| Length | 153 |
| Max. P | 0.997272 |

| Location | 21,326,575 – 21,326,673 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.69 |
| Mean single sequence MFE | -21.62 |
| Consensus MFE | -13.48 |
| Energy contribution | -14.28 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21326575 98 - 27905053 UCAGUUUGCAAACAUGCAUUUGGCCAAAAGCCAACAGGGCCAACAAAAAAAGG------AGCACAGUG------UGGAAAAUGCUGUGACAACGAUAGUCGACUAUCAGC ......((((....)))).((((((............))))))........(.------.((((((((------(.....)))))))).)..)(((((....)))))... ( -25.40) >DroSec_CAF1 18917 92 - 1 UCAGUUUGCAAACAUGCAUUUGGCCAAAAGCCAACAGGGCCAACAAAAAAAGG------AGCACGCCG------------ACAGUGUGAAAGCGAUAGUCGACUAUCAGA ...(((((((....)))..((((((............))))))..........------..(((((..------------...))))).))))(((((....)))))... ( -21.30) >DroSim_CAF1 18044 92 - 1 UCAGUUUGCAAACAUGCAUUUGGCCAAAAGCCAACAGGGCCAACAAAAAAAGG------AGCACGCCG------------ACAGUGUGAAAGCGAUAGUCGACUAUCAGA ...(((((((....)))..((((((............))))))..........------..(((((..------------...))))).))))(((((....)))))... ( -21.30) >DroEre_CAF1 19114 80 - 1 UCAGUUUGCAAACAUGCAUUUGGCCAACAGCCAACGGGGCCAACAAAAAA------------------------UCAAACAAGCUGUGA------CGUUCGACUAUCAGA .(((((((((....)))..((((((............)))))).......------------------------......))))))...------............... ( -14.60) >DroYak_CAF1 17887 110 - 1 UCAGUUUGCAAACAUGCAUUUGGCCAAAAGCCAACAGGGCCAACAAAAAAAAGAAAACGAACACGCCGACAGUGUGGAAAAAGCUGUGAAAACGAUAGUCGACUAUCAGA ...((((....(((.((..((((((............))))))..................(((((.....)))))......)))))..))))(((((....)))))... ( -25.50) >consensus UCAGUUUGCAAACAUGCAUUUGGCCAAAAGCCAACAGGGCCAACAAAAAAAGG______AGCACGCCG______U__AA_AAGCUGUGAAAACGAUAGUCGACUAUCAGA .((((.((((....)))).((((((............)))))).......................................)))).......(((((....)))))... (-13.48 = -14.28 + 0.80)



| Location | 21,326,633 – 21,326,728 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.98 |
| Mean single sequence MFE | -36.63 |
| Consensus MFE | -14.34 |
| Energy contribution | -14.37 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.39 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21326633 95 + 27905053 GCCCUGUUGGCUUUUGGCCAAAUGCAUGUUUGCAAACUGAAG------C---GGCUGACAAUUAACUAAUGUCAGCCGCACAAAGUUCAG-------UUUGUAGAUGUUGG ((....(((((.....)))))..))(((((((((((((((((------(---((((((((.........))))))))))......)))))-------)))))))))))... ( -42.20) >DroVir_CAF1 13931 101 + 1 GCCGCGUUGGCUUUUGGCCAACUGCAUGUUUGCUAACGGACAAACCUCC---GGCUGACAAUUAACAAAUGUCCACAGCGCACAACUUGC-------GCCAAGGAUGCGGG .((((((..((..(((((.(((.....))).)))))((((......)))---)))..))...........((((...(((((.....)))-------))...)))))))). ( -37.50) >DroGri_CAF1 14516 111 + 1 GCCGUAUUGGCUUUUGGCCAACUGCUUGUUUGCAAACCAAAAACCCCCACAAGGCUGACAAUUAACAAAUGUCAACGGCACAGAAGCUUCCCCAAAAGCCAAGGAUGCGGG .((((((((((((((((......((((.(.(((.............((....)).(((((.........)))))...))).).))))....))))))))))...)))))). ( -34.50) >DroEre_CAF1 19154 95 + 1 GCCCCGUUGGCUGUUGGCCAAAUGCAUGUUUGCAAACUGAAG------C---GGCUGACAAUUAACUAAUGUCAGCCGCACAAAGUUUCG-------CUUGGAGAUGUGGG ..(((.(((((.....)))))..((((.((.(((((((...(------(---((((((((.........))))))))))....))))).)-------)...)).))))))) ( -38.30) >DroYak_CAF1 17957 95 + 1 GCCCUGUUGGCUUUUGGCCAAAUGCAUGUUUGCAAACUGAAG------C---GGCUGACAAUUAACUAAUGUCAGCCGCACAAAGUUUCG-------UUCGGAGAUGUGGG .(((..(((((.....)))))..((((.((((.(((((...(------(---((((((((.........))))))))))....)))))..-------..)))).))))))) ( -36.60) >DroMoj_CAF1 13381 101 + 1 GCCGUGUUGGCUUUUGGCCAACUGCAUGUUUGCUAGCCGAAAACCUCCA---AGCUGACAAUUAACAAAUGUCAACGGUGCAAAACUUGU-------GCCAAAGAUGCGGG .(((((((((.(((((((.....(((....)))..))))))).))....---.(.(((((.........))))).)((..((.....)).-------.))...))))))). ( -30.70) >consensus GCCCUGUUGGCUUUUGGCCAAAUGCAUGUUUGCAAACUGAAA______C___GGCUGACAAUUAACAAAUGUCAACCGCACAAAAUUUCG_______GCCAAAGAUGCGGG (((...(((((.....)))))..(((....)))...................)))(((((.........)))))....((((...(((.............))).)))).. (-14.34 = -14.37 + 0.03)



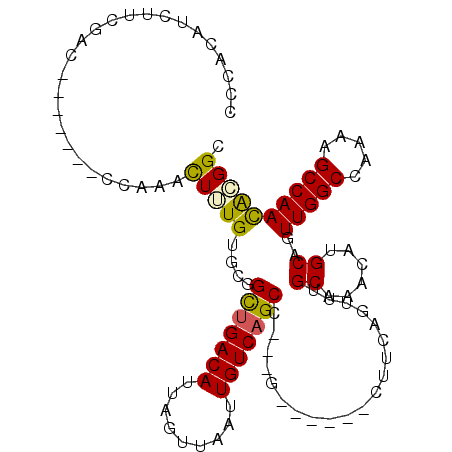
| Location | 21,326,633 – 21,326,728 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.98 |
| Mean single sequence MFE | -35.47 |
| Consensus MFE | -16.29 |
| Energy contribution | -15.77 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21326633 95 - 27905053 CCAACAUCUACAAA-------CUGAACUUUGUGCGGCUGACAUUAGUUAAUUGUCAGCC---G------CUUCAGUUUGCAAACAUGCAUUUGGCCAAAAGCCAACAGGGC ((........((((-------(((((......((((((((((.........))))))))---)------))))))))))...........(((((.....)))))..)).. ( -34.90) >DroVir_CAF1 13931 101 - 1 CCCGCAUCCUUGGC-------GCAAGUUGUGCGCUGUGGACAUUUGUUAAUUGUCAGCC---GGAGGUUUGUCCGUUAGCAAACAUGCAGUUGGCCAAAAGCCAACGCGGC .((((.(((.((((-------(((.....))))))).)))..(((((((((.(.(((((---...)).))).).)))))))))......((((((.....)))))))))). ( -38.20) >DroGri_CAF1 14516 111 - 1 CCCGCAUCCUUGGCUUUUGGGGAAGCUUCUGUGCCGUUGACAUUUGUUAAUUGUCAGCCUUGUGGGGGUUUUUGGUUUGCAAACAAGCAGUUGGCCAAAAGCCAAUACGGC ...((.((((..(...)..)))).))......((((((((((.........)))))))...(((..((((((((((((((......))))...))))))))))..)))))) ( -40.20) >DroEre_CAF1 19154 95 - 1 CCCACAUCUCCAAG-------CGAAACUUUGUGCGGCUGACAUUAGUUAAUUGUCAGCC---G------CUUCAGUUUGCAAACAUGCAUUUGGCCAACAGCCAACGGGGC (((..........(-------(.(((((....((((((((((.........))))))))---)------)...)))))))..........(((((.....))))).))).. ( -35.70) >DroYak_CAF1 17957 95 - 1 CCCACAUCUCCGAA-------CGAAACUUUGUGCGGCUGACAUUAGUUAAUUGUCAGCC---G------CUUCAGUUUGCAAACAUGCAUUUGGCCAAAAGCCAACAGGGC (((...........-------..(((((....((((((((((.........))))))))---)------)...)))))(((....)))..(((((.....)))))..))). ( -33.20) >DroMoj_CAF1 13381 101 - 1 CCCGCAUCUUUGGC-------ACAAGUUUUGCACCGUUGACAUUUGUUAAUUGUCAGCU---UGGAGGUUUUCGGCUAGCAAACAUGCAGUUGGCCAAAAGCCAACACGGC ...(((.(((....-------..)))...))).(((((((((.........))))))).---((..((((((.(((((((.........))))))).))))))..)).)). ( -30.60) >consensus CCCACAUCUUCGAC_______CCAAACUUUGUGCGGCUGACAUUAGUUAAUUGUCAGCC___G______CUUCAGUUUGCAAACAUGCAGUUGGCCAAAAGCCAACACGGC ..........................(((((....(((((((.........)))))))....................((......))..(((((.....)))))))))). (-16.29 = -15.77 + -0.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:47 2006