
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,303,225 – 21,303,400 |
| Length | 175 |
| Max. P | 0.933984 |

| Location | 21,303,225 – 21,303,324 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.03 |
| Mean single sequence MFE | -47.57 |
| Consensus MFE | -22.26 |
| Energy contribution | -23.27 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21303225 99 + 27905053 ---GGCGGAAUGCCA---CCCAUGGGAGGUA---CACCGCCGCGACCGGGAAUGAUGCCCGGAAUGCCUCCGGGUGCAAUGCAACCCGGCGGACCCAUGCAGCCCAGU ---(((((..((((.---((....)).))))---..)))))(((.(((((.......)))))....((.((((((((...)).)))))).)).....)))........ ( -46.40) >DroVir_CAF1 4935 102 + 1 CCUACGGGCAUGCAG---CCUAUGGGCGGCC---CCGGACCGAGACCCGGCUUAAUGAGCGGCAUGCCACCGGGCGGAAUGCCGUCCGUAGGUCAACUGCCUGCAGGA (((.((((((....(---(((((((((((((---((((.(((.....)))..........((....)).)))))......)))))))))))))....)))))).))). ( -51.90) >DroPse_CAF1 30899 103 + 1 -----CGGCAUGCAACAGCCCAUGGGUGGUGGGGUGCCACCACGUCCUGGCCUGAUGCCGGGCAUGCCACCAGGUGGCAUGUCGCCCGUGGGCCCUAUGCCACCGGGA -----.(((((((............(((((((....)))))))...(((((.....)))))))))))).((.(((((((((..(((....)))..))))))))).)). ( -59.30) >DroSec_CAF1 31846 99 + 1 ---GGCGGAAUGCCA---CCCAUGGGCGGUA---CGCCGCCGCGACCAGGAAUGAUGCCCGGAAUGCCUCCGGGUGCAAUGCAACCCGGCGGACCCAUGCAGCCCAGU ---(((((..((((.---((....)).))))---..)))))(((....((........))((....((.((((((((...)).)))))).))..)).)))........ ( -42.50) >DroEre_CAF1 50655 99 + 1 ---GGCGGAAUGCCA---CCCAUGGGCGGUG---CUCCGCCGCGACCAGGAAUGAUGCCCGGAAUGCCACCGGGCGCAAUGCAACCCGGCGGACCCAUGCAACCCAGU ---((((((..(((.---((....)).))).---.)))))).((.((.((......((((((.......))))))((...))..)).))))................. ( -41.20) >DroYak_CAF1 33283 99 + 1 ---GGCGGAAUGCCA---CCCAUGGGCGGUA---CUCCGCCGCGACCAGGAAUGAUGCCCGGAAUGCCACCGGGUGCAAUGCAGCCCGGCGGACCCAUGCAACCCAGU ---((((((.((((.---((....)).))))---.))))))(((....((........))((....((.((((((((...))).))))).))..)).)))........ ( -44.10) >consensus ___GGCGGAAUGCCA___CCCAUGGGCGGUA___CGCCGCCGCGACCAGGAAUGAUGCCCGGAAUGCCACCGGGUGCAAUGCAACCCGGCGGACCCAUGCAACCCAGU ....................((((((((((........)))))..((.((.......)).))....((.((((((........)))))).))..)))))......... (-22.26 = -23.27 + 1.00)



| Location | 21,303,225 – 21,303,324 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.03 |
| Mean single sequence MFE | -50.38 |
| Consensus MFE | -23.75 |
| Energy contribution | -25.67 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21303225 99 - 27905053 ACUGGGCUGCAUGGGUCCGCCGGGUUGCAUUGCACCCGGAGGCAUUCCGGGCAUCAUUCCCGGUCGCGGCGGUG---UACCUCCCAUGGG---UGGCAUUCCGCC--- ((((((.(((.((((.((.((((((.((...)))))))).))...)))).))).....))))))...(((((.(---(.((.((....))---.)).)).)))))--- ( -45.10) >DroVir_CAF1 4935 102 - 1 UCCUGCAGGCAGUUGACCUACGGACGGCAUUCCGCCCGGUGGCAUGCCGCUCAUUAAGCCGGGUCUCGGUCCGG---GGCCGCCCAUAGG---CUGCAUGCCCGUAGG .(((((.((((((.(.((((.((.((((.....((((((((((.....)))......)))))))((((...)))---))))).)).))))---).)).)))).))))) ( -46.30) >DroPse_CAF1 30899 103 - 1 UCCCGGUGGCAUAGGGCCCACGGGCGACAUGCCACCUGGUGGCAUGCCCGGCAUCAGGCCAGGACGUGGUGGCACCCCACCACCCAUGGGCUGUUGCAUGCCG----- .((.((((((((.(.(((....)))..))))))))).)).(((((((.......(((.(((((..(((((((....))))))))).))).)))..))))))).----- ( -56.30) >DroSec_CAF1 31846 99 - 1 ACUGGGCUGCAUGGGUCCGCCGGGUUGCAUUGCACCCGGAGGCAUUCCGGGCAUCAUUCCUGGUCGCGGCGGCG---UACCGCCCAUGGG---UGGCAUUCCGCC--- ((..((.(((.((((.((.((((((.((...)))))))).))...)))).))).....))..))...(((((.(---(.(((((....))---))).)).)))))--- ( -46.10) >DroEre_CAF1 50655 99 - 1 ACUGGGUUGCAUGGGUCCGCCGGGUUGCAUUGCGCCCGGUGGCAUUCCGGGCAUCAUUCCUGGUCGCGGCGGAG---CACCGCCCAUGGG---UGGCAUUCCGCC--- ((..((.(((.((((.(((((((((.((...)))))))))))...)))).))).....))..))...(((((((---..(((((....))---)))..)))))))--- ( -53.10) >DroYak_CAF1 33283 99 - 1 ACUGGGUUGCAUGGGUCCGCCGGGCUGCAUUGCACCCGGUGGCAUUCCGGGCAUCAUUCCUGGUCGCGGCGGAG---UACCGCCCAUGGG---UGGCAUUCCGCC--- ((..((.(((.((((.((((((((.(((...)))))))))))...)))).))).....))..))...(((((((---(.(((((....))---))).))))))))--- ( -55.40) >consensus ACUGGGUUGCAUGGGUCCGCCGGGCUGCAUUGCACCCGGUGGCAUUCCGGGCAUCAUUCCUGGUCGCGGCGGAG___UACCGCCCAUGGG___UGGCAUUCCGCC___ ...(((.(((....(.((.((((((........)))))).)))...(((((.......)))))....(((((.......)))))...........))).)))...... (-23.75 = -25.67 + 1.92)


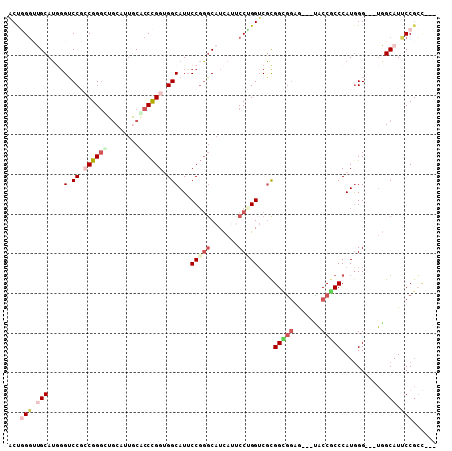
| Location | 21,303,253 – 21,303,361 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.40 |
| Mean single sequence MFE | -55.60 |
| Consensus MFE | -32.59 |
| Energy contribution | -32.92 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21303253 108 - 27905053 GGAGCUGCUGCAUCUGCUGUGCCUGCAGCAUGU---GUGGACUGGGCUGCAUGGGUCCGCCGGGUUGCAUUGCACCCGGAGGCAUUCCGGGCAUCAUUCCCGGUCGCGGCG ...(((((.((((.((((((....)))))).))---)).(((((((.(((.((((.((.((((((.((...)))))))).))...)))).))).....)))))))))))). ( -54.60) >DroVir_CAF1 4966 111 - 1 GCAGCUGCUGCAUUUGCUGCGCCUGCAGUAAAUGCUGUUGUCCUGCAGGCAGUUGACCUACGGACGGCAUUCCGCCCGGUGGCAUGCCGCUCAUUAAGCCGGGUCUCGGUC .(((((((((((((((((((....)))))))))))(((......)))))))))))......(..((((((.((((...)))).))))))..).....((((.....)))). ( -48.60) >DroPse_CAF1 30931 111 - 1 GGAGCUGCUGCAUUUGCUGGGCCUGCAGCAUAUGCUGUUGUCCCGGUGGCAUAGGGCCCACGGGCGACAUGCCACCUGGUGGCAUGCCCGGCAUCAGGCCAGGACGUGGUG .......(((....((((((((..(((((....)))))((((((((((((((.(.(((....)))..))))))))).)).)))).)))))))).)))((((.....)))). ( -53.60) >DroEre_CAF1 50683 111 - 1 GGAGCUGCUGCAUCUGCUGCGCCUGCAGCAUGUGCUGUGGACUGGGUUGCAUGGGUCCGCCGGGUUGCAUUGCGCCCGGUGGCAUUCCGGGCAUCAUUCCUGGUCGCGGCG ...(((((.((((.((((((....)))))).))))....(((..((.(((.((((.(((((((((.((...)))))))))))...)))).))).....))..)))))))). ( -59.80) >DroYak_CAF1 33311 111 - 1 GGAGCUGCUGCAUUUGCUGCGCCUGCAGCAUGUGCUGUGGACUGGGUUGCAUGGGUCCGCCGGGCUGCAUUGCACCCGGUGGCAUUCCGGGCAUCAUUCCUGGUCGCGGCG ...(((((.((((.((((((....)))))).))))....(((..((.(((.((((.((((((((.(((...)))))))))))...)))).))).....))..)))))))). ( -60.20) >DroPer_CAF1 39532 111 - 1 GGAGCUGCUGCAUUUGCUGGGCCUGCAGCAUAUGCUGUUGUCCCGGUGGCAUAGGGCCCACGGGCGACAUGCCACCUGGUGGCAUGCCCGGCAUUAGGCCAGGGCGUGGUG ((..(.((.((((.(((((......))))).)))).)).)..))((((((((.(.(((....)))..)))))))))......((((((((((.....))).)))))))... ( -56.80) >consensus GGAGCUGCUGCAUUUGCUGCGCCUGCAGCAUAUGCUGUGGACCGGGUGGCAUGGGUCCGACGGGCGGCAUUCCACCCGGUGGCAUGCCGGGCAUCAUGCCAGGUCGCGGCG ...(((((.((((.((((((....)))))).))))....(((((((........((((..((((..........))))..((....))))))......)))))))))))). (-32.59 = -32.92 + 0.34)


| Location | 21,303,290 – 21,303,400 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.31 |
| Mean single sequence MFE | -46.23 |
| Consensus MFE | -26.15 |
| Energy contribution | -25.43 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21303290 110 - 27905053 UCC-UGUUUCAUUUGGGUGGCAUUUGUCUGCUUAUCAUGCGGAGCUGCUGCAUCUGCUGUGCCUGCAGCAUGU---GUGGACUGGGCUGCAUGGGUCCGCCGGGUUGCAUUGCA .((-((.......))))..(((..(((..((((..(((((((..(..(..(((.((((((....)))))).))---)..)...)..)))))))((....)))))).))).))). ( -36.70) >DroVir_CAF1 5003 105 - 1 ------AAUCACUUGGGCGGCAG---UUUGCCCAUCAUGCGCAGCUGCUGCAUUUGCUGCGCCUGCAGUAAAUGCUGUUGUCCUGCAGGCAGUUGACCUACGGACGGCAUUCCG ------........(((.(((((---((.((.(.....).)))))))))(((((((((((....)))))))))))((((((((.(.((((....).))).))))))))).))). ( -41.90) >DroPse_CAF1 30968 109 - 1 -CA-CUCCUCACUUGGGCGGCAG---CUUGCCCAUCAUGCGGAGCUGCUGCAUUUGCUGGGCCUGCAGCAUAUGCUGUUGUCCCGGUGGCAUAGGGCCCACGGGCGACAUGCCA -..-..((......))(((((((---(((((.......)).))))))))))....((((((.(.(((((....))))).).))))))(((((.(.(((....)))..)))))). ( -51.10) >DroSim_CAF1 31961 113 - 1 UCC-CGGUUCAUUUGGGUGGCAUUUGUCUGCCCAUCAUGCGGAGCUGCUGCAUCUGCUGUGCCUGCAGCAUGUGCUGUGGACUGGGCUGCAUGGGUCCGCCGGGUUGCAUUGCA .((-((((.....((((((((....))).)))))(((((((((((..(.((((.((((((....)))))).)))).)..).))...))))))))....)))))).......... ( -48.90) >DroYak_CAF1 33348 114 - 1 UCCCCAGUUCAUUUGGGUGGCAUUUGUCUGCCCAUCAUGCGGAGCUGCUGCAUUUGCUGCGCCUGCAGCAUGUGCUGUGGACUGGGUUGCAUGGGUCCGCCGGGCUGCAUUGCA ..(((((.....)))))..(((..(((..((((..((((((((((..(.((((.((((((....)))))).)))).)..).))...)))))))((....)))))).))).))). ( -49.00) >DroPer_CAF1 39569 109 - 1 -CA-CUUCUCACUUGGGCGGCAG---CUUGCCCAUCAUGCGGAGCUGCUGCAUUUGCUGGGCCUGCAGCAUAUGCUGUUGUCCCGGUGGCAUAGGGCCCACGGGCGACAUGCCA -..-............(((((((---(((((.......)).))))))))))....((((((.(.(((((....))))).).))))))(((((.(.(((....)))..)))))). ( -49.80) >consensus _CC_CGGUUCACUUGGGCGGCAG___UCUGCCCAUCAUGCGGAGCUGCUGCAUUUGCUGCGCCUGCAGCAUAUGCUGUGGACCGGGUGGCAUGGGUCCGACGGGCGGCAUUCCA .............(((((((.......)))))))..((((...(((((.((((.((((((....)))))).)))).)).((((.(......).)))).....))).)))).... (-26.15 = -25.43 + -0.72)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:36 2006