
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,264,360 – 21,264,452 |
| Length | 92 |
| Max. P | 0.970786 |

| Location | 21,264,360 – 21,264,452 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 91.03 |
| Mean single sequence MFE | -16.58 |
| Consensus MFE | -14.66 |
| Energy contribution | -15.22 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
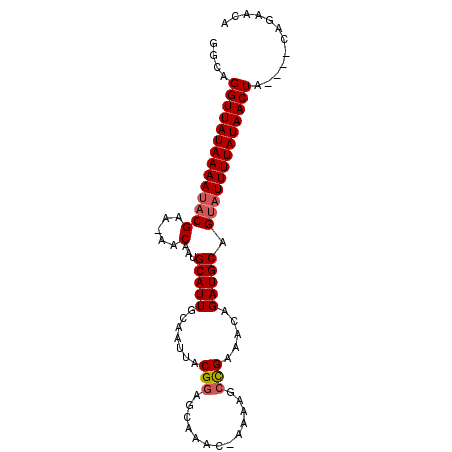
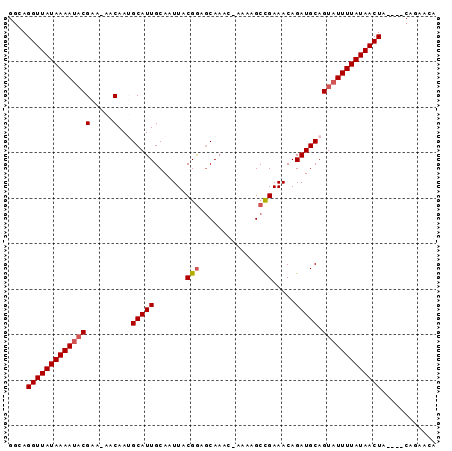
>3R_DroMel_CAF1 21264360 92 + 27905053 GGCAGGUUAUAAAAUACGAA-AACAAGGCAUUGCAAUUACGGAGCAAAC-AAAAGCCGAAACAGAUGCAGUAUUUUAUAACUAACAGCGGAACA .((.(((((((((((((...-......(((((.......(((.......-.....))).....))))).)))))))))))))....))...... ( -18.90) >DroSec_CAF1 89251 88 + 1 GGCAGGUUAUAAAAUACGAA-AACAAUGCAUUGCAAUUACGGAUCAAAC-ACAAGCUGAAACAGAUGCAGUAUUUUAUAACUA----CUGAACA ..((((((((((((((((..-..)..((((((.......(((.......-.....))).....))))))))))))))))))..----))).... ( -17.00) >DroSim_CAF1 92006 89 + 1 GGCAGGUUAUAAAAUACGAA-AACAAUGCAUUGCAAUUACGGAGCAAAAAAAAAGCCGAAACAGAUGCAGUAUUUUAUAACUA----CAGAACA ....((((((((((((((..-..)..((((((.......(((.............))).....))))))))))))))))))).----....... ( -19.02) >DroYak_CAF1 98762 89 + 1 GGCAGGUUAUAAAACUCGAAAAACAAAGCAUUGCAAUUACGAAGCAAAC-AAAAGCCGAAACAGAUGCAGUAUUUUAUAACUA----CAGAACA ....((((((((((..((.....)...(((((.......((..((....-....)))).....))))).)..)))))))))).----....... ( -11.40) >consensus GGCAGGUUAUAAAAUACGAA_AACAAUGCAUUGCAAUUACGGAGCAAAC_AAAAGCCGAAACAGAUGCAGUAUUUUAUAACUA____CAGAACA ....((((((((((((((.....)...(((((.......(((.............))).....))))).)))))))))))))............ (-14.66 = -15.22 + 0.56)

| Location | 21,264,360 – 21,264,452 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 91.03 |
| Mean single sequence MFE | -17.61 |
| Consensus MFE | -13.25 |
| Energy contribution | -13.31 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
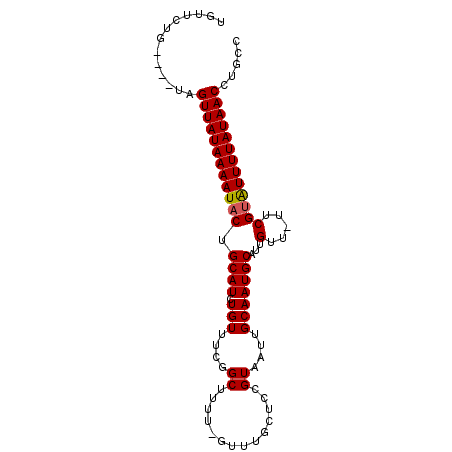
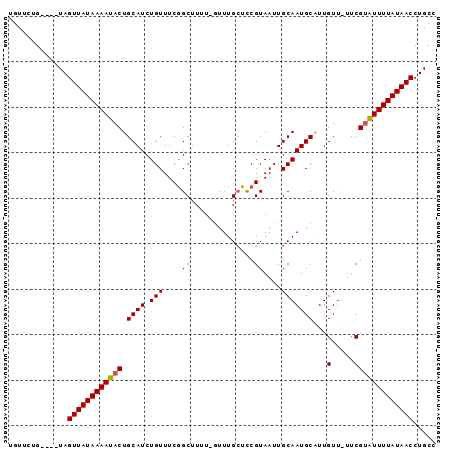
>3R_DroMel_CAF1 21264360 92 - 27905053 UGUUCCGCUGUUAGUUAUAAAAUACUGCAUCUGUUUCGGCUUUU-GUUUGCUCCGUAAUUGCAAUGCCUUGUU-UUCGUAUUUUAUAACCUGCC .........((..((((((((((((.((....))...(((..((-((((((...))))..)))).))).....-...))))))))))))..)). ( -18.30) >DroSec_CAF1 89251 88 - 1 UGUUCAG----UAGUUAUAAAAUACUGCAUCUGUUUCAGCUUGU-GUUUGAUCCGUAAUUGCAAUGCAUUGUU-UUCGUAUUUUAUAACCUGCC ......(----((((((((((((((((((..((..(((((....-).))))..))....)))).......(..-..)))))))))))).)))). ( -17.50) >DroSim_CAF1 92006 89 - 1 UGUUCUG----UAGUUAUAAAAUACUGCAUCUGUUUCGGCUUUUUUUUUGCUCCGUAAUUGCAAUGCAUUGUU-UUCGUAUUUUAUAACCUGCC ......(----(((((((((((((((((((.(((..(((.............))).....)))))))).....-...))))))))))).)))). ( -18.32) >DroYak_CAF1 98762 89 - 1 UGUUCUG----UAGUUAUAAAAUACUGCAUCUGUUUCGGCUUUU-GUUUGCUUCGUAAUUGCAAUGCUUUGUUUUUCGAGUUUUAUAACCUGCC .....((----((((........))))))........(((..((-((..(((.((.((..((((....))))..)))))))...))))...))) ( -16.30) >consensus UGUUCUG____UAGUUAUAAAAUACUGCAUCUGUUUCGGCUUUU_GUUUGCUCCGUAAUUGCAAUGCAUUGUU_UUCGUAUUUUAUAACCUGCC .............((((((((((((.((((.(((....((..............))....)))))))...(.....)))))))))))))..... (-13.25 = -13.31 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:29 2006