
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,259,177 – 21,259,268 |
| Length | 91 |
| Max. P | 0.932413 |

| Location | 21,259,177 – 21,259,268 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.99 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -14.50 |
| Energy contribution | -14.26 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
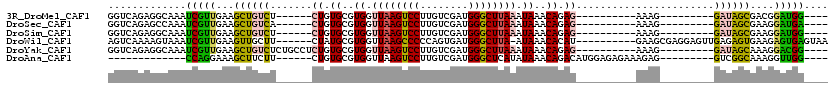
>3R_DroMel_CAF1 21259177 91 + 27905053 GGUCAGAGGCAAAUCGUUGAAGCUGUCU------CUGUGCGUGGUUAAGUCCUUGUCGAUGGGCUUAAAUAAACAGAG----------AAAG---------GAUAGCGACGGAUGG---- .((.....))...((((((...((.(((------((((...((.((((((((........)))))))).)).))))))----------)..)---------)....))))))....---- ( -27.70) >DroSec_CAF1 84046 91 + 1 GGUCAGAGCCAAAUCGUUGAAGCUGUCA------CUGUGCGUGGUUAAGUCCUUGUCGAUGGGCUUAAAUAAACAGAG----------AAAG---------GAUAGCGAAGGAUGA---- (((....)))...(((((...((((((.------((.((..((.((((((((........)))))))).))..)).))----------....---------))))))....)))))---- ( -26.40) >DroSim_CAF1 86770 91 + 1 GGUCAGAGGCAAAUCGUUGAAGCUGUCU------CUGUGCGUGGUUAAGUCCUUGUCGAUGGGCUUAAAUAAACAGAG----------AAAG---------GAUAGCGAAGGAUGG---- .((.....))...((((((...((.(((------((((...((.((((((((........)))))))).)).))))))----------).))---------..)))))).......---- ( -26.80) >DroWil_CAF1 86603 103 + 1 AGUCAAAAGUAAAUCGUUGAAGUUGCUU------CUAUGCGUGGUUAAGCCCCCAGUGAUGGGCUUA-AUAAACACAU----------GAAGCGAGGAGUUGAGAGUGAAGAGUGAGUAA (.(((........((.(..(..((((((------(.(((.((.(((((((((........)))))))-))..)).)))----------)))))))....)..)))........))).).. ( -27.39) >DroYak_CAF1 93553 97 + 1 GGUCAGAGGCAAAUCGUUGAAGCUGUCUCUGCCUCUGUGCGUGGUUAAGUCCUUGUCGAUGGGCUUAAAUAAACAGAG----------AAAG---------GAUAGCAAAGGACGG---- .(((.((......))......((((((.((..((((((...((.((((((((........)))))))).)).))))))----------..))---------))))))....)))..---- ( -33.60) >DroAna_CAF1 77837 88 + 1 -------------CCAGGAAAGCUUCUU------CUGUGCGUGGUUAAGUCCUUGUCGAUGGGCUCAUAUAAACAGACAUGGAGAGAAAGAG---------GUCGGCAAAGGUUGG---- -------------........(((((((------((.(.((((.((((((((........)))))....))).....)))).).)).)))))---------))((((....)))).---- ( -20.00) >consensus GGUCAGAGGCAAAUCGUUGAAGCUGUCU______CUGUGCGUGGUUAAGUCCUUGUCGAUGGGCUUAAAUAAACAGAG__________AAAG_________GAUAGCGAAGGAUGG____ .............(((((...((((((.......((.((..((.((((((((........)))))))).))..)).)).......................))))))....))))).... (-14.50 = -14.26 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:24 2006