| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,256,797 – 21,256,933 |
| Length | 136 |
| Max. P | 0.990413 |

| Location | 21,256,797 – 21,256,895 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -17.72 |
| Energy contribution | -17.84 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21256797 98 + 27905053 CGCUUAUUGUUCCGUUGCCGGAACUUUGUUUACAUUUGCCAUCAUUUUCUGGUGGCAUAACAAAAAUAAUUUUUGCUCUUCAGUCAUCGUAGCAGCUC .(((((((((((((....))))))((((((......((((((((.....)))))))).)))))))))))....(((((..........).)))))).. ( -24.70) >DroSec_CAF1 81785 88 + 1 CGCUUAUUGUUCCGUUGCCGGAACUUUGUUUACACAUGCCAUCAUUUUCUGGUGGCAUAACAAAAAUAAUUUUUGCUCUACUGCCAU----------C .((.....((((((....))))))((((((.....(((((((((.....)))))))))))))))..........))...........----------. ( -23.40) >DroSim_CAF1 84509 98 + 1 CGCUUAUUGUUCCGUUGCCGGAACUUUGUUUACAUUUGCCAUCAUUUUCUGGUGGCAUAACAAAAAUAAUUUUUGCUCUACAGCCAUCGUAGCAGCUC ...(((((((((((....))))))((((((......((((((((.....)))))))).))))))))))).....(((((((.......)))).))).. ( -27.90) >DroYak_CAF1 91336 98 + 1 CGCUUAUUGUUCUGCUGCCGGAACUUUGUUUACAUUUGCCAUCACUUUCUGGUGGCAUAAUAAAAAUAAUUUUUGCUCUGCAGUCAUCGCAGCAGCUC ........((((((....)))))).((((((..(((((((((((.....)))))))).)))..)))))).....(((((((.......)))).))).. ( -26.00) >DroAna_CAF1 75821 94 + 1 UGCUUAUUGUCCUGUCGCCGG-GCUUUGUUUACACACACAAUCACUUU-UGGUGGCAGGACGA-AAUAAU-UUCGCUUUACAGUCGUCUUGGCAGCUC .(((..(((((((((((((((-(..((((........)))).....))-))))))))))))))-......-...(((..((....))...)))))).. ( -28.90) >consensus CGCUUAUUGUUCCGUUGCCGGAACUUUGUUUACAUUUGCCAUCAUUUUCUGGUGGCAUAACAAAAAUAAUUUUUGCUCUACAGUCAUCGUAGCAGCUC .((.....((((((....))))))((((((......((((((((.....)))))))).))))))..........))...................... (-17.72 = -17.84 + 0.12)

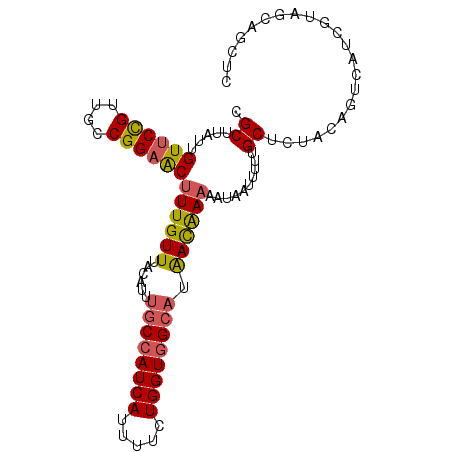

| Location | 21,256,797 – 21,256,895 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -15.78 |
| Energy contribution | -16.46 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21256797 98 - 27905053 GAGCUGCUACGAUGACUGAAGAGCAAAAAUUAUUUUUGUUAUGCCACCAGAAAAUGAUGGCAAAUGUAAACAAAGUUCCGGCAACGGAACAAUAAGCG ..(((..((((..........(((((((.....))))))).(((((.((.....)).)))))..))))......((((((....))))))....))). ( -28.20) >DroSec_CAF1 81785 88 - 1 G----------AUGGCAGUAGAGCAAAAAUUAUUUUUGUUAUGCCACCAGAAAAUGAUGGCAUGUGUAAACAAAGUUCCGGCAACGGAACAAUAAGCG (----------.(((((....(((((((.....))))))).))))).)...........((.(((....)))..((((((....)))))).....)). ( -27.10) >DroSim_CAF1 84509 98 - 1 GAGCUGCUACGAUGGCUGUAGAGCAAAAAUUAUUUUUGUUAUGCCACCAGAAAAUGAUGGCAAAUGUAAACAAAGUUCCGGCAACGGAACAAUAAGCG ..(((.(((((.....)))))(((((((.....))))))).(((((.((.....)).)))))............((((((....))))))....))). ( -29.30) >DroYak_CAF1 91336 98 - 1 GAGCUGCUGCGAUGACUGCAGAGCAAAAAUUAUUUUUAUUAUGCCACCAGAAAGUGAUGGCAAAUGUAAACAAAGUUCCGGCAGCAGAACAAUAAGCG (..(((((((...((((((...))..........(((((..(((((.((.....)).)))))...)))))...))))...)))))))..)........ ( -23.20) >DroAna_CAF1 75821 94 - 1 GAGCUGCCAAGACGACUGUAAAGCGAA-AUUAUU-UCGUCCUGCCACCA-AAAGUGAUUGUGUGUGUAAACAAAGC-CCGGCGACAGGACAAUAAGCA ..(((.......((.((....))))..-......-..((((((.(.((.-...((..((((........)))).))-..)).).))))))....))). ( -20.20) >consensus GAGCUGCUACGAUGACUGUAGAGCAAAAAUUAUUUUUGUUAUGCCACCAGAAAAUGAUGGCAAAUGUAAACAAAGUUCCGGCAACGGAACAAUAAGCG ......................((.................(((((.((.....)).)))))............((((((....)))))).....)). (-15.78 = -16.46 + 0.68)

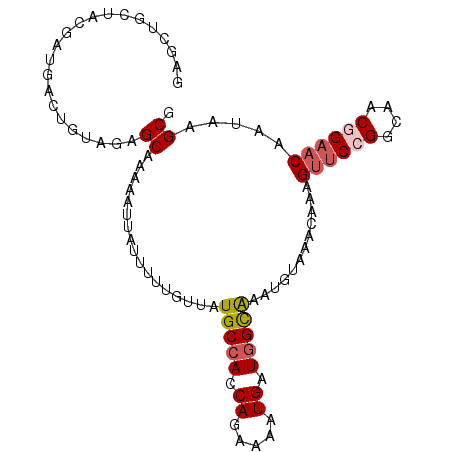
| Location | 21,256,827 – 21,256,933 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.96 |
| Mean single sequence MFE | -23.22 |
| Consensus MFE | -13.42 |
| Energy contribution | -14.70 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21256827 106 + 27905053 UACAUUUGCCAUCAUUUUCUGGUGGCAUAACAAAAAUAAUUUUUGCUCUUCAGUCAUCGUAGCAGCUCAUCA--AAGAUAGUUGUAUAAGGCAUUUAUGCAAGCCAAA (((...((((((((.....))))))))...(((((.....))))).............)))((((((.(((.--..)))))))))....(((..........)))... ( -21.70) >DroSec_CAF1 81815 96 + 1 UACACAUGCCAUCAUUUUCUGGUGGCAUAACAAAAAUAAUUUUUGCUCUACUGCCAU----------CAUCA--AAGAUAGUUGAAGAAGGCAUUUAUGCAAGCCAAA .....(((((((((.....))))))))).............(((((.....((((.(----------(.(((--(......)))).)).)))).....)))))..... ( -26.20) >DroSim_CAF1 84539 106 + 1 UACAUUUGCCAUCAUUUUCUGGUGGCAUAACAAAAAUAAUUUUUGCUCUACAGCCAUCGUAGCAGCUCAUCA--AAGAUAGUUGAACAAGGCAUUUAUGCAAGCCAAA ......((((((((.....))))))))..............(((((......(((...((..(((((.(((.--..)))))))).))..)))......)))))..... ( -22.90) >DroYak_CAF1 91366 102 + 1 UACAUUUGCCAUCACUUUCUGGUGGCAUAAUAAAAAUAAUUUUUGCUCUGCAGUCAUCGCAGCAGCUCAUCA--AAGAUAGUUGAACAAGGCAUU----CAAGCCAAA ......((((((((.....)))))))).................(((((((.......)))).)))...(((--(......))))....(((...----...)))... ( -24.90) >DroAna_CAF1 75850 96 + 1 UACACACACAAUCACUUU-UGGUGGCAGGACGA-AAUAAU-UUCGCUUUACAGUCGUCUUGGCAGCUCAUCAAAAAGAUAGUUGAACAGGGCAUUUAUC--------- ....(((.(((......)-))))).((((((((-..(((.-......)))...))))))))...((((.((((........))))...)))).......--------- ( -20.40) >consensus UACAUUUGCCAUCAUUUUCUGGUGGCAUAACAAAAAUAAUUUUUGCUCUACAGUCAUCGUAGCAGCUCAUCA__AAGAUAGUUGAACAAGGCAUUUAUGCAAGCCAAA ......((((((((.....))))))))...(((((.....))))).......(((.......(((((.(((.....)))))))).....)))................ (-13.42 = -14.70 + 1.28)

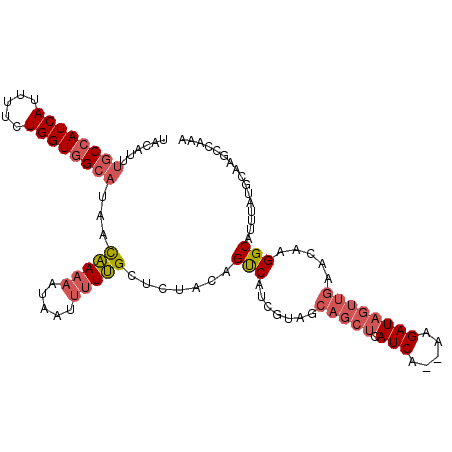
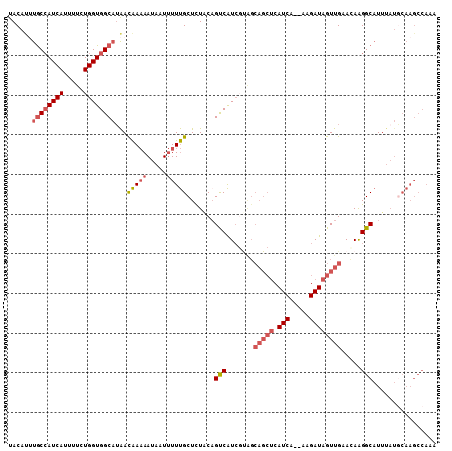
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:58:20 2006